Abstract
We have investigated the spatial relationship between transcription sites and chromosome territories in the interphase nucleus of human female fibroblasts. Immunolabeling of nascent RNA was combined with visualization of chromosome territories by fluorescent in situ hybridization (FISH). Transcription sites were found scattered throughout the territory of one of the two X chromosomes, most likely the active X chromosome, and that of both territories of chromosome 19. The other X chromosome territory, probably the inactive X chromosome, was devoid of transcription sites. A distinct substructure was observed in interphase chromosome territories. Intensely labeled subchromosomal domains are surrounded by less strongly labeled areas. The intensely labeled domains had a diameter in the range of 300–450 nm and were sometimes interconnected, forming thread-like structures. Similar large scale chromatin structures were observed in HeLa cells expressing green fluorescent protein (GFP)-tagged histone H2B. Strikingly, nascent RNA was almost exclusively found in the interchromatin areas in chromosome territories and in between strongly GFP-labeled chromatin domains. These observations support a model in which transcriptionally active chromatin in chromosome territories is markedly compartmentalized. Active loci are located predominantly at or near the surface of compact chromatin domains, depositing newly synthesized RNA directly into the interchromatin space.
Keywords: chromosome territory, transcription sites, nuclear organization, chromatin structure, fluorescent in situ hybridization
The dynamic structure of the interphase nucleus is an important factor in the control of gene expression. Many nuclear functions, such as replication, transcription, and RNA processing, occupy well-defined domains in the interphase nucleus (Spector 1993; Wansink et al. 1994b; Van Driel et al. 1995; De Jong et al. 1996; Jackson 1997; Lamond and Earnshaw 1998). Mammalian chromosomes in the interphase nucleus are localized in distinct territories (Lichter et al. 1988; Pinkel et al. 1988; Cremer et al. 1993). The spatial organization of nuclear domains with respect to chromosome territories has become a focus of interest in the analysis of functional architecture of the interphase nucleus. For instance, Zirbel et al. 1993 have shown that components of the splicing machinery, as well as specific gene transcripts of an integrated human papilloma virus genome, are often excluded from the interior of chromosome territories. Furthermore, several coding genes were found positioned preferentially at the periphery of chromosome territories. Apparently, this configuration was independent of the transcriptional activity of the genes. Noncoding genomic DNA was found more randomly positioned in chromosome territories (Kurz et al. 1996). Since these observations are related to only a small set of genomic sequences, further studies are needed to explore whether nuclear compartmentalization with respect to the structural organization of interphase chromosomes is of functional significance for gene control.
The aim of the present study is to elucidate whether transcription sites have a distinct spatial distribution in relation to the organization of chromosome territories and chromatin. Detection of nascent RNA with the use of 5-bromouridine 5′-triphosphate (BrUTP)1 by in vivo and in vitro labeling procedures was developed by Wansink et al. 1993 and Jackson et al. 1993. These studies have shown that transcription sites in cultured mammalian cells are concentrated in numerous domains that occur scattered throughout the nucleoplasm. In the present study, detection of nascent RNA was combined with labeling of chromosome territories by fluorescence in situ hybridization (FISH), using chromosome-specific DNA whole library probes. Labeling of nascent RNA in combination with visualization of X chromosome and chromosome 19 territories was analyzed in diploid human female fibroblasts. In mammals, one X chromosome is transcriptionally inactivated during early development, while the other remains active (Lyon 1961; Gartler and Goldman 1994; Rastan 1994; Heard et al. 1997). The two X chromosome territories in somatic female cell nuclei provide an excellent model system to study the distribution of transcription sites in the active X chromosome (Xa), using the inactive X chromosome (Xi) as an internal control.
Our results show that transcription sites are scattered throughout the chromosome territories of Xa and chromosome 19, whereas Xi is devoid of transcription sites. We observe that interphase chromosome territories show a distinct substructure, consisting of domains that were intensely labeled in the FISH procedure, surrounded by less intensely labeled areas. Strikingly, newly synthesized and nascent RNA was preferentially present in interchromatin areas within chromosome territories. These interchromatin areas contain little or no DNA. Since in situ hybridization involves relatively harsh conditions, we also analyzed the relationship between chromatin and transcription sites by investigating the spatial relationship between GFP-tagged histone H2B and transcription sites. Nascent RNA was present in between intensely labeled chromatin domains, similar to that observed after performing FISH. These results indicate that there is a marked compartmentalization of transcriptionally active and inactive chromatin in chromosome territories.
Materials and Methods
Cell Culture
Human primary fibroblasts (kindly provided by Dr. L.H.F. Mullenders, University of Leiden, The Netherlands) with a normal female karyotype (46, XX) were grown at 37°C under a 2.5% CO2 atmosphere in Ham's F-10 (GIBCO BRL), supplemented with 15% (wt/vol) heat inactivated FCS (Boehringer Mannheim Corp.), 2 mM l-glutamine (GIBCO BRL), 100 IU/ml penicillin and 100 μg/ml streptomycin (GIBCO BRL). For BrUTP labeling, as well as in situ hybridization, cells were cultured on Alcian blue coated coverslips (Brink et al. 1992). Cells were grown to 50–70% confluency before use.
HeLa cells were stably transfected with a H2B–GFP vector (kindly provided by Dr. G. Wahl, Salk Institute for Biological Studies, La Jolla, CA; Kanda et al. 1998) by Dr. M. Kimura (Sir William Dunn School of Pathology, Oxford, UK). Cells were grown at 37°C under 10% CO2 atmosphere in DME, supplemented with 10% FCS, and were kept continuously under 0.35 mg/ml G418 (Sigma Chemical Co.) drug selection. To visualize H2B–GFP in living cells, cells were grown on glass-bottom microwell dishes coated with poly-d-lysine (Mattek Co.).
Labeling of Nascent RNA
Run-on labeling nascent RNA in permeabilized cells by incorporation of BrUTP has been described in detail by Wansink et al. 1993, Wansink et al. 1994a. In brief, cells were detergent-permeabilized and incubated with a run-on transcription buffer, containing 0.5 mM ATP, CTP, GTP, and BrUTP for 20 min at room temperature. BrUTP incorporated into nascent RNA was visualized by indirect immunofluorescent labeling. Cells were fixed for 15 min at 4°C in 2% (wt/vol) formaldehyde, diluted in PBS. After fixation, cells were permeabilized with 0.5% (wt/vol) Triton X-100 (Sigma Chemical Co.) in PBS for 5 min and incubated with PBS containing 100 mM glycine (Sigma Chemical Co.) for 10 min. Subsequently, cells were incubated overnight at 4°C with a rat mAb raised against BrdU (Seralab) diluted 1:500 in PBS, containing 0.1 μg/ml herring sperm DNA to block nonspecific binding. After several washes in PBS, cells were incubated for 1 h at room temperature with either Cy3-conjugated donkey anti–rat antibody (Jackson ImmunoResearch Laboratories, Inc.), or FITC-conjugated donkey anti–rat antibody (Jackson ImmunoResearch Laboratories, Inc.), diluted in PBS containing 0.1 μg/ml herring sperm DNA. Next, cells were washed in PBS and used for the in situ hybridization procedure. Control experiments, using UTP instead of BrUTP, inhibition by α-amanitin, and RNase-dependent destruction of BrU-containing RNA, have been described previously (Wansink et al. 1993, Wansink et al. 1994a).
In vivo labeling of nascent RNA was carried out as described by Wansink et al. 1993, Wansink et al. 1994a. In brief, living cells were microinjected into the cytoplasm with 100 mM BrUTP in 140 mM KCl and 2 mM piperazine-N,N′-bis (2-ethane sulfonic acid), pH 7.4. Approximately 5% cell volume was injected. After microinjection, cells were cultured for 10 min at 37°C and subsequently fixed and labeled as described above.
Immunofluorescent Labeling of Acetylated Histone H4, Centromeres and PML Bodies
For immunofluorescent labeling, cells were fixed for 10 min at 4°C in 2% (wt/vol) formaldehyde in PBS. After fixation, cells were permeabilized with 0.5% (wt/vol) Triton X-100 in PBS for 5 min and incubated with PBS containing 100 mM glycine for 10 min. Subsequently, cells were incubated for 1 h at 37°C with an antibody recognizing acetylated histone H4, an antibody recognizing centromeres, or an antibody recognizing PML. Rabbit polyclonal antibodies (R232 and R252) against acetylated histone H4 (kindly provided by Dr. B. Turner, University of Birmingham, UK) were used. R232 polyclonal antibody recognizes histone H4 acetylated at lysine 8, and antibody R252 histone H4 acetylated at lysine 16 (Turner and Fellows 1989; Turner et al. 1989). Human autoimmune serum H33 recognizes centromeres (kindly provided by Dr. W.J. Van Venrooy, Katholieke Universiteit Nijmegen, The Netherlands). To label PML protein in PML antibodies, we used the mouse mAb 5E10 (Stuurman et al. 1992; Koken et al. 1994). R232 and R252 antibodies were diluted 1:1,000, anticentromere 1:50,000, and 5E10 1:3 in PBS containing 0.1 μg/ml herring sperm DNA. After several washes in PBS, cells were incubated with appropriate secondary antibodies, using either Cy3- or FITC-conjugated donkey anti–rabbit antibody, goat anti–human antibody, or donkey anti–mouse antibody (Jackson ImmunoResearch Laboratories, Inc.). Secondary antibodies were diluted in PBS containing 0.1 μg/ml herring sperm DNA and incubations were performed for 1 h at room temperature. As a control, cells were incubated in the absence of primary antibody. Subsequently, cells were washed in PBS and used for the in situ hybridization procedure.
Chromosome-specific Probes
Painting of the human X chromosome and of chromosome 19 was achieved using chromosome-specific biotin-labeled DNA library probes (Cambio). Probes were denatured at 85°C for 5 min. To suppress labeling of repetitive sequences, which are often not chromosome-specific, denatured probe samples (85°C, 5 min) were self-annealed (2–3 μg/ml, 60–100 min, 37°C) before use. Preannealed probes gave the same result in the FISH procedures as using a large excess of Cot-1 DNA. The probes have been shown to label their respective metaphase chromosomes completely and specifically, showing the specificity of the DNA library probes (Carter et al. 1990; Telenius et al. 1992). In some experiments, we specifically labeled only repetitive sequences. In that case, we used human Cot-1 DNA that was directly conjugated to FITC or Cy3 (kindly provided by Dr. J.C.A.G. Wiegant, University of Leiden, The Netherlands; 20 μg/ml).
In Situ Hybridization Procedure
The FISH procedure was an adaptation of protocols described by Lichter et al. 1988, Cremer et al. 1988, Cremer et al. 1993, and Kurz et al. 1996. Fixation, pretreatment, and denaturation procedures were subjected to several modifications. The following protocol was found to produce optimal preservation of intranuclear organization of transcription sites and H4 acetylated histones. Cells were rinsed with PBS and fixed for 10 min at 4°C in 4% (wt/vol) formaldehyde diluted in PBS. To facilitate probe penetration, cells were treated with 0.1 M HCl for 10 min and subsequently with a mixture of 0.5% Triton and 0.5% (wt/vol) saponin in PBS for 10 min. After washing in PBS, genomic DNA was denatured by incubating cells in 2× SSC containing 70% formamide at 73°C for 4 min, followed by incubation in 2× SSC containing 50% formamide for 1 min. Hybridization with the biotin tagged probe was allowed to proceed overnight at 42°C. Posthybridization washes with 2× SSC containing 50% formamide and subsequent washes with 2× SSC were performed at 42°C. After blocking in 4× SSC, containing 5% (wt/vol) nonfat dry milk (NFDM; Coberco) for 30 min at room temperature, cells were incubated with FITC or Cy3-conjugated streptavidin, rinsed, and incubated with biotin-conjugated antibody from goat against avidin (Vector Labs, Inc.), subsequently rinsed and incubated once more with FITC or Cy3-conjugated streptavidin. All incubations for fluorescent labeling were performed at room temperature during 20 min in 4× SSC containing 5% (wt/vol) NFDM and 0.1 μg/ml herring sperm DNA. The wash steps after antibody incubations were performed with 4× SSC, 0.05% Tween-20. After fluorescent labeling, DNA staining was performed with 0.4 μg/ml Hoechst 33258 (Sigma Chemical Co.) in PBS, or with 0.5 μM Sytox green (Molecular Probes) in 25 mM Hepes buffer, pH 7.4. As a control, cells were incubated in hybridization buffer in the absence of probe. Slides were mounted in Vectashield (Brunschwig). Slides were kept at 4°C until evaluation and analyzed within 24 h.
Confocal Scanning Laser Microscopy
Images were recorded with a Leica or LSM 510 Zeiss confocal laser scanning microscope both equipped with a 100×/1.23 NA oil immersion lens. With the Leica microscope, a dual-wavelength argon ion laser was used, whereas the Zeiss confocal microscope used an argon laser at 488 nm in combination with a helium neon laser at 543 nm to excite green and red fluorochromes simultaneously at 488 nm and 514 nm, respectively. Emitted fluorescence was detected in the Leica confocal microscope using a 525 DF10 bandpass filter for FITC and a 550-nm longpass filter for Cy3, whereas in the Zeiss microscope a 505-530-bandpass filter for green signal and a 560-nm longpass filter for the red signal was used. Pairs of images were collected simultaneously in the green and red channels. Three dimensional (3-D) images were scanned as 512 × 512 × 32 voxel images (sampling rate 49-nm lateral and 208-nm axial).
Image Processing
Images were corrected for optical cross-talk (Manders et al. 1992). Image analysis was performed using SCIL-IMAGE software (Ten Kate et al. 1990; Van Balen et al. 1994). Images were subjected to a 3-D image restoration procedure to correct for diffraction-induced distortion, using the Huygens System 2 (Scientific Volume Imaging BV; Van der Voort and Straster 1985). The image restoration procedure uses a measured point spread function, which was obtained at precisely the same conditions as the image. To this end, 200-nm fluorescent beads (FluoSpheres; Molecular Probes, Inc.) were imaged. The 3-D image restoration procedure significantly improved the quality of the 3-D images by removing Poisson noise and by deblurring the images.
Results
Structural Preservation during In Situ Hybridization
We have developed a modified chromosome painting protocol with improved signal to background ratio, based on the chromosome painting procedures described by Lichter et al. 1988, Cremer (1993), and Kurz et al. 1996. It was important to establish that the relatively harsh FISH procedure did not alter nuclear structure in general, and chromosome structure in particular. To investigate the degree of structural preservation, we have carried out four tests. First, we analyzed whether the FISH procedure induced an altered spatial distribution of transcription sites. Second, we investigated the effect of the FISH procedure on the distribution of DNA stained with Sytox green. Third, we analyzed the spatial distribution of acetylated histone H4 before and after the chromosome painting procedure. Histone acetylation is generally correlated with gene activity and reduced acetylation levels with gene silencing (Hebbes et al., 1988; Braunstein et al. 1993). Fourth, we measured in one and the same cell nucleus before and after the FISH procedure the spatial distribution of centromeres and of PML bodies (Stuurman et al. 1992).
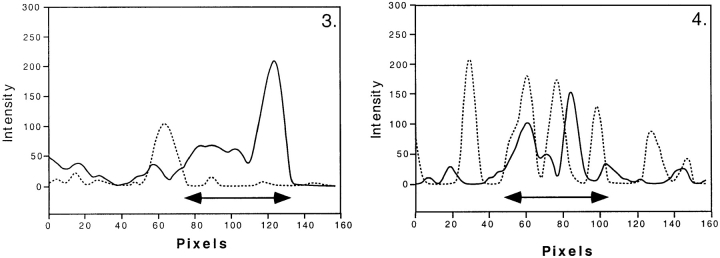
Visual inspection shows that the spatial distribution of nascent RNA did not change in any recognizable way due to the FISH procedure (Fig. 1A and Fig. B). Staining of DNA with Sytox green with or without chromosome painting showed that the FISH procedure also did not induce major changes in DNA distribution (Fig. 1C and Fig. D). After the FISH procedure, the DNA staining pattern seemed slightly more blurred and the overall DNA labeling appeared more intense. Fig. 1E and Fig. F, show immunofluorescent labeling of histone H4 acetylated at lysine 8, before and after FISH labeling. The punctate distribution of acetylated H4 throughout the nucleoplasm did not significantly change after chromosome painting. Only the diffuse, low intensity component in the labeling was somewhat diminished after FISH, making the intense punctate granular labeling more prominent. This may be due to extraction of some histone protein during FISH labeling. These results show that the FISH procedure does not result in major rearrangements in DNA, chromatin, and nascent RNA.
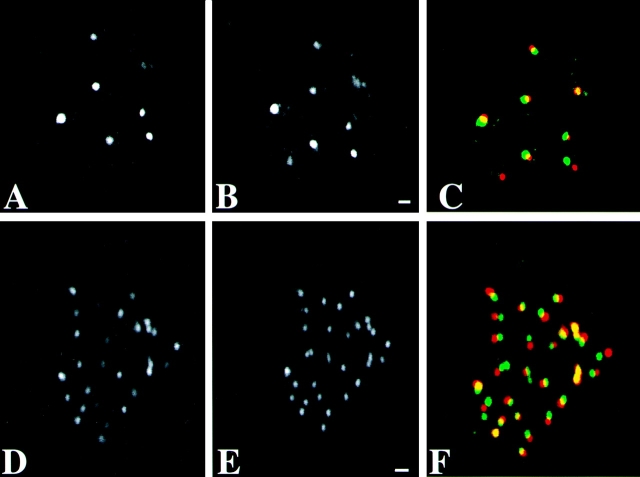
Figure 1.
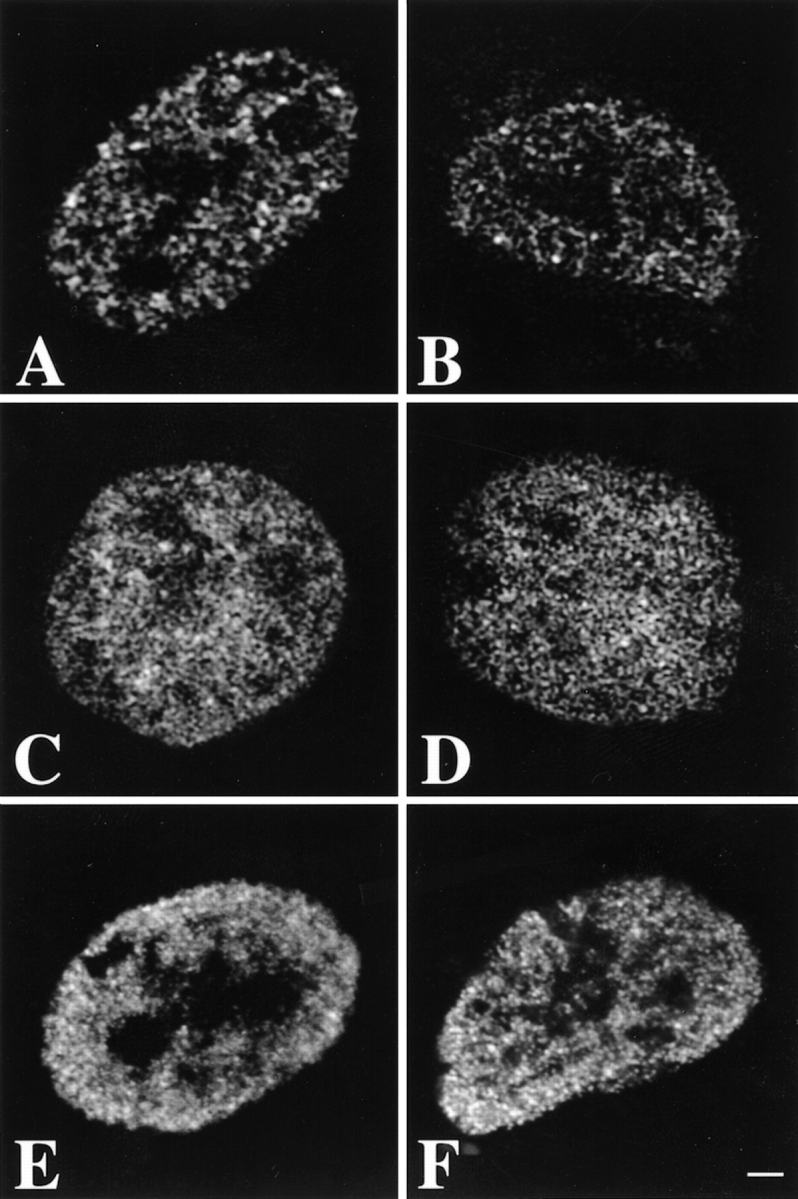
Preservation of the spatial distribution of nascent RNA, DNA, and acetylated histone H4 in human primary fibroblasts during the FISH procedure. Optical sections were obtained before (A, C, and E) and after (B, D, and F) carrying out the FISH protocol. A and B, Optical sections of nuclei in which transcription sites were immunofluorescently labeled. The distribution of nascent RNA did not change significantly during the FISH procedure. C and D, Optical sections of nuclei labeled with the fluorescent DNA stain Sytox green. The pattern of DNA staining did not change significantly after in situ hybridization. E and F, Optical sections of nuclei in which acetylated histone H4 was fluorescently labeled. The distribution did not change significantly during the chromosome painting procedure. The diffuse, low intensity labeling observed before FISH was diminished after the procedure, resulting in a somewhat more pronounced granular labeling. Images shown have been subjected to 3-D image restoration. Bar, 1.75 μm.
Fig. 2 shows the spatial distribution of PML bodies (Fig. 2, A–C) and of centromeres (Fig. 2, D–F) imaged in the same cell before and after FISH. The corresponding optical sections before and after the FISH procedure (PML bodies, Fig. 2A and Fig. B, and centromeres, D and E, respectively) show that nuclear organization is well-preserved during the FISH procedure. Both the integrity and the spatial distribution of the nuclear bodies and of the centromeres are essentially unchanged. The small changes in spatial distribution (see overlay in Fig. 2C and Fig. F) can be attributed largely to a slight tilting of the nucleus relative to the substratum, due to the FISH procedure.
Figure 2.
Preservation of the spatial distribution of centromeres and PML bodies in human primary fibroblasts during the FISH procedure. The spatial distribution of PML bodies and of centromeres were analyzed before and after the FISH procedure in the same nucleus. A, B, and C, Corresponding individual optical sections of the same nucleus labeled with anti-PML antibody are shown in A (before the FISH procedure) and B (after FISH). C shows an overlay of A (red) and B (green). Bar, 0.84 μm. D, E, and F, Corresponding individual optical sections of the same nucleus labeled with anticentromere antibody. D, Before FISH procedure. E, After FISH procedure. F shows an overlay of D (red) and E (green). The FISH procedure results in only small changes in the distribution of PML bodies and centromeres. These changes are in part due to a slight tilting of the nuclei during FISH labeling. Bar, 0.84 μm.
Together, these results establish that the FISH procedure does not induce significant changes in the spatial distribution of subnuclear structures at the light microscopy level. In particular, we did not observe any collapse, aggregation, unfolding, or other major rearrangement of chromatin in the nucleus. We conclude that, at the resolution level of the light microscope, nuclear structure is not affected by the chromosome painting procedure.
Substructure of Chromosome Territories
A striking feature that emerges from our confocal images after FISH labeling is that chromosome territories display a distinct substructure (Fig. 3). Chromosome territories do not appear as compact objects. Rather, they show a modulated intensity distribution inside the territory. Since the chromosome-specific library-probes label the complete metaphase chromosome, it is unlikely that considerable parts of the interphase chromosome remain unlabeled (Carter et al. 1990; Telenius et al. 1992). The chromosome territories contained strongly labeled chromosomal subdomains, surrounded by less intensely labeled areas. The strongly labeled subchromosomal structures have a diameter in the range of 300–450 nm (Fig. 3 and Fig. 4). Intensely labeled parts of a chromosome often seemed interconnected, forming thread-like, folded structures. A similar, distinct substructure in nuclei stained for DNA with Sytox green was observed (Fig. 1C and Fig. D) and in nuclei of HeLa cells expressing GFP-histone H2B (see Visualization of Chromatin Using GFP-histone H2B; Fig. 5), suggesting a reticular organization of chromatin. It is tempting to suggest that, at least locally, the chromatin fiber that constitutes the chromosome can be followed in an optical section.
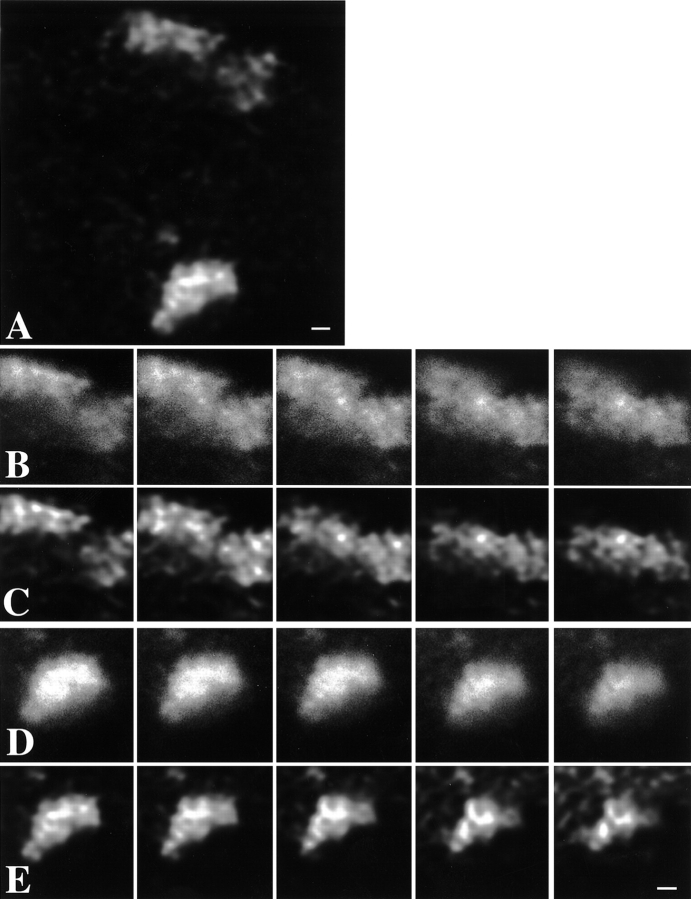
Figure 3.
Chromosome territories have a distinct substructure. X chromosome territories in nuclei of female primary fibroblasts were labeled by FISH, using a chromosome-specific DNA probe library. A, Single optical section through the center of a nucleus showing two labeled X chromosome territories. The image has undergone 3-D image restoration. Bar, 2.1 μm. C and E, Five consecutive optical sections (step size along z-axis is 0.2 μm) showing structural details of the two territories visible in A. The images shown have been subjected to 3-D image restoration. Territories show strongly labeled chromosomal subdomains surrounded by less intensely labeled areas. Intensely labeled chromosomal subdomains have a diameter in the range of 300–450 nm. In several cases, subchromosomal domains appear interconnected, forming thread-like structures (also see Fig. 4). Bar, 4.2 μm. B and D, Same optical sections as in C and E, respectively. Shown are unprocessed, crude images that have not undergone 3-D image restoration. Structural details that are enhanced after 3-D image restoration are visible in the unprocessed optical sections. Bar, 4.2 μm.
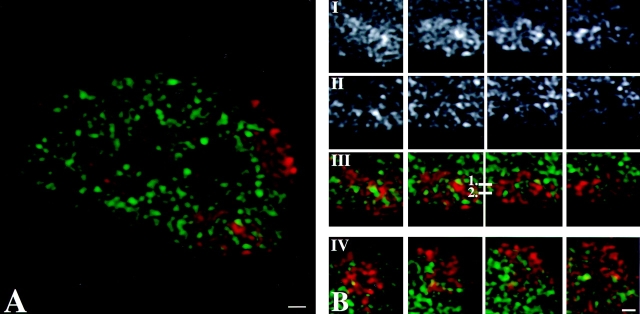
Figure 4.
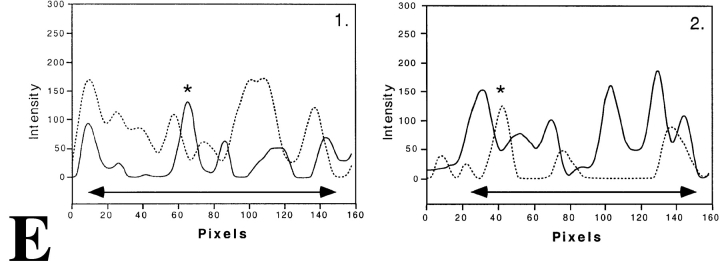
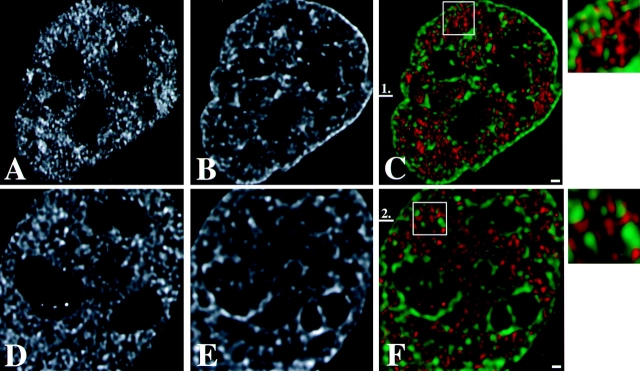
Distribution of transcription sites in relation to territories of the two X chromosomes and chromosome 19 in female primary fibroblasts. Labeling of nascent RNA by incorporation of BrUTP (green) in permeabilized cells (run-on-transcription) is combined with FISH labeling of X chromosome territories (A–C) and of chromosome 19 territories (D). In all nuclei analyzed, transcription sites occur as defined spots throughout one of the two X chromosome territories (most likely Xa; B) and in both chromosome 19 territories (D). Almost no transcription sites are observed in the other X chromosome territory (most likely Xi; C). Nascent RNA preferentially accumulates between the chromosomal subdomains in the areas with little or no FISH label. A, Single optical section through the center of a cell nucleus with labeled Xa and Xi chromosome territories (red) and transcription sites (green). Bar, 0.875 μm. B, Four consecutive optical sections (z-distance 0.2 μm) through an X chromosome territory that contains transcription sites (most likely Xa). I and II represent consecutive optical sections of the X chromosome territory and of transcription sites, respectively. III shows the overlay of the chromosome territory (red) and the transcription sites (green). IV shows in another nucleus an X-chromosome territory (red) containing transcription sites (green). Bar, 0.66 μm. C, Four consecutive optical sections (z-distance 0.2 μm) through a single X chromosome territory not containing transcription sites (most likely Xi). I and II represent consecutive optical sections of the X chromosome territory and transcription sites, respectively. III shows the overlay of the chromosome territory (red) and the transcription sites (green). IV shows in another nucleus an X-chromosome territory (red) that does not contain transcription sites (green). Bar, 0.66 μm. D, Four consecutive optical sections (z-distance 0.2 μm) through a single chromosome 19 territory. I and II represent consecutive optical sections of the chromosome 19 territory and transcription sites, respectively. III shows the overlay of the chromosome territory (red) and the transcription sites (green), respectively. IV shows a chromosome 19 territory (red) and transcription sites (green) in another nucleus. Bar, 0.66 μm. All images shown have undergone 3-D image restoration. Bar, 0.66 μm. E, Line scans through individual Xa, Xi and chromosome 19 territories. The dotted curve represents the local intensity of the nascent RNA signal, the continuous line shows the intensity distribution of the FISH-labeled chromosome. Line scans are shown through the Xa (1 and 2), Xi (3), and chromosome 19 (4) territories. The position of each line scan is indicated by a horizontal bar with a number in B and C (chromosome Xa and Xi, respectively) and D (chromosome 19). Transcription sites with their center of gravity precisely on the line are marked with an asterisk in 1 and 2. The two-arrow horizontal line below the intensity distributions marks the size of the chromosome territory.
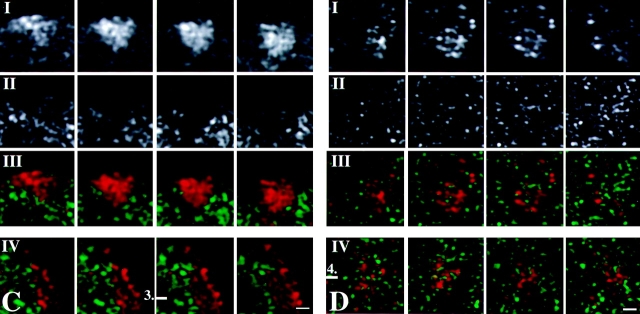
Figure 5.
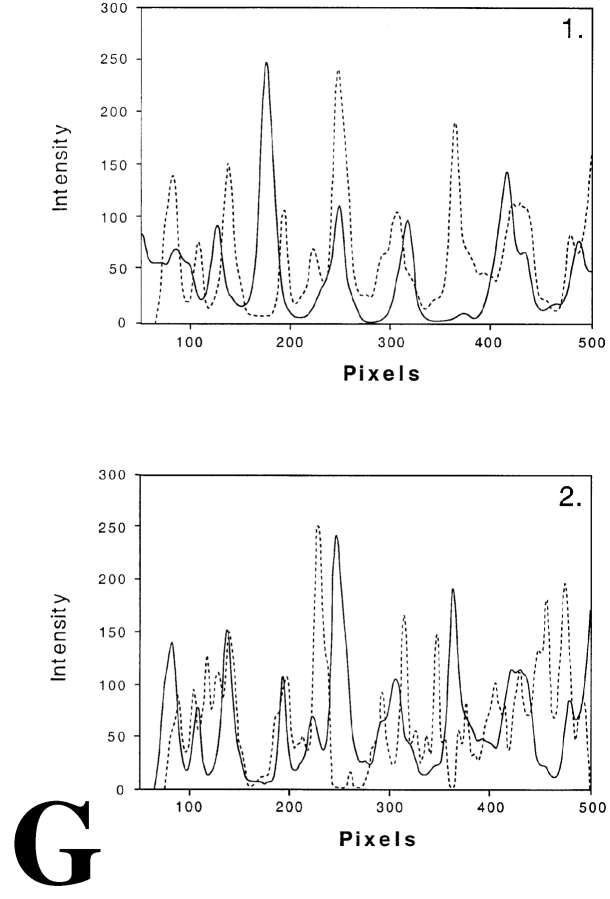
Distribution of transcription sites in relation to chromatin in HeLa cells expressing GFP tagged histone H2B. Transcription sites were visualized after in vivo incorporation of BrUTP into nascent RNA. Nascent RNA accumulated preferentially in nuclear domains that contain little or no GFP-labeled chromatin. A and D, Single optical sections of two different nuclei showing the distribution of transcription sites. B and E, Same optical as shown in A and D, respectively, showing the distribution of GFP-histone H2B. C and F, Overlay of A and B, and D and E, respectively, showing the spatial relationship between transcription sites (red) and chromatin domains (green). Magnified areas in C and F (white squares) are shown in the inserts at the right. The images show that nascent RNA is predominantly found in areas that contain little or no GFP-labeled chromatin. All images shown are subjected to 3-D image restoration. G, Line scans through compact GFP-histone H2B domains and transcription sites. The dotted curve represents the local intensity of the nascent RNA signal, the continuous line shows the intensity distribution of the GFP-histone H2B signal. The position of the line scan is indicated by a horizontal bar with a number in C (1) and F (2). Bars, 0.6 μm.
Images were subjected to 3-D image restoration to correct for diffraction-induced distortions. The advantage of such image restoration is that it significantly improves the quality of the 3-D images. It is conceivable that the restoration procedure preferentially enhances certain spatial frequencies. To verify this, we compared the unprocessed individual consecutive sections through a chromosome territory (Fig. 3B and Fig. D) to the same sections after 3-D image restoration (Fig. 3C and Fig. E). Careful comparison shows that essentially all details of a chromosome territory in the processed image are precisely matched by features that can be found in the preprocessed image. Apparent slight differences are most likely due to the fact that the 3-D restored images contain information from optical sections above and below those that are shown, whereas the unprocessed images do not. Very low intensity signal may be lost by the image restoration procedure, probably because it cannot be discriminated from noise in the restoration process. These results show that no artificial structures in chromosome territories are created by the 3-D restoration algorithm.
To investigate whether the less intensely labeled areas in chromosome territories contained repetitive DNA, which is suppressed during FISH labeling, we performed hybridization with a probe specific for highly repetitive human Cot-1 DNA, in dual labeling with an X chromosome probe (data not shown). Hybridization of metaphase chromosomes with the Cot-1 probe revealed intense labeling predominantly in the centromeric region, which is known to contain a high concentration of repetitive DNA. Simultaneous in situ hybridization with the X chromosome probe and the probe recognizing highly repetitive DNA showed that repetitive DNA does not occur concentrated in the areas in the territory that are less intensely labeled with the chromosome-specific probe in the FISH procedure. We conclude that the less intensely labeled areas in chromosome territories constitute a compartment that contains little or no DNA.
Transcription Sites throughout Chromosome Territories
To establish the 3-D distribution of transcription sites in relation to chromosome territories, chromosome painting was combined with the visualization of transcription sites. Fluorescent labeling of nascent RNA shows that transcription occurs scattered throughout the nucleoplasm (Jackson et al. 1993; Wansink et al. 1993, Wansink et al. 1994a), often with exception of nucleoli, due to inaccessibility of the anti-BrU antibody (Wansink et al. 1993). The 3-D images show that transcription sites occur throughout the territory of one of two X chromosomes (Fig. 4A and Fig. B), whereas almost no transcription sites are found in the other X chromosome (Fig. 4A and Fig. C). Most likely, the X chromosome territory containing transcription sites represents the Xa and the territory devoid of transcription is the Xi. We also analyzed the distribution of transcription sites with respect to the autosomal chromosome 19. Chromosome 19 replicates early in S phase and is relatively gene rich, as indicated by the number of CpG islands (Craig and Bickmore 1994). Transcription sites were found throughout the chromosome 19 territories (Fig. 4 D), similar to that observed for Xa. These results show that transcription sites are present throughout chromosome territories with exception of the Xi chromosome territory.
Importantly, no striking differences in the substructure of the territories of the Xa, Xi, or chromosome 19 can be seen. This implies that the substructure does not depend on differences in overall transcriptional activity, or on differences between sex chromosomes and autosomal chromosomes.
Relationship between Transcription Sites and Chromosomal Substructure
We have analyzed the relationship between the spatial distribution of transcription sites and the substructure of chromosome territories. Strikingly, transcription sites (green signal in Fig. 4 A, and B III and IV) in Xa, as well as in chromosome 19 (Fig. 4 D, III and IV), were mainly found between the intensely labeled chromosomal subdomains (red signal in Fig. 4 A, B III and IV, and D III and IV) and almost never overlapped with them. Line scans were made to obtain more quantitative information about the spatial relationship between transcription sites and chromosome substructure. Fig. 4 E shows typical examples of line scans through Xa (Fig. 4 E, 1 and 2) and Xi (Fig. 4 E, 3) and chromosome 19 (Fig. 4 E, 4) territories. The position of the scans are marked in Fig. 4B III and C IV. The line scans through the territory of putative Xa and of chromosome 19 territories confirm that transcription sites (indicated by arrowheads) are often localized near the surface of the intensely labeled chromosomal subdomains (Fig. 4 E, 1–3). The transcription sites marked with an asterisk have their 3-D center of gravity exactly on the scanned line. These transcription sites clearly show that nascent and newly synthesized RNA accumulates near a chromosomal subdomain, generally not overlapping with it.
Visualization of Chromatin Using GFP-histone H2B
Despite our efforts (see Structural Preservation during In Situ Hybridization) we cannot fully rule out that the FISH procedure induces local rearrangement of chromatin and/or nascent RNA. Therefore, we have used an alternative procedure to label chromatin, using cells that stably express GFP-tagged histone H2B (Kanda et al. 1998). Since essentially all histone protein is incorporated in chromatin, the distribution of GFP–H2B faithfully represents that of chromatin (Kanda et al. 1998). Obviously, this procedure does not allow visualization of individual chromosome territories. However, it does allow analysis of the chromatin distribution in nuclei under in vivo conditions. Intense GFP-labeling is seen particularly at the nuclear periphery and around nucleoli, suggesting a compact, heterochromatin-like local structure. In addition, an irregular, apparently reticular labeling of chromatin is seen throughout the nucleoplasm, with exception of the nucleoli. It is important to note that, like after FISH labeling, intensely labeled areas are observed with a diameter in the range of 300–450 nm (Fig. 5B and Fig. E). Visualizing transcription sites in cells expressing GFP–H2B showed strikingly little overlap between transcription sites (red signal in Fig. 5C, Fig. F, and both enlargements) and chromatin (green signal in Fig. 5C, Fig. F, and both enlargements). Line scans confirm these observations (Fig. 5 G, 1 and 2). This observation is fully in agreement with the results after FISH labeling of chromatin in chromosome territories (see Relationship between Transcription Sites and Chromosomal Substructure). These results confirm that FISH does not affect nuclear structure at the light microscopy level and underscore our observations showing that chromosome territories have a distinct substructure and that transcription sites are located near the surface of intensely labeled subchromosomal domains.
Discussion
Two major principles of organization of the interphase nucleus can be distinguished. First, many nuclear components and processes occur compartmentalized in well-defined domains in the interphase nucleus (Spector et al., 1993; Wansink et al. 1994b; Van Driel et al. 1995; De Jong et al. 1996; Lamond and Earnshaw 1998; Mistelli and Spector, 1998). Second, individual chromosomes occupy discrete chromosome territories that are apparently not intruded by chromatin from other chromosomes (Lichter et al. 1988; Pinkel et al. 1988; Cremer et al. 1993). The dynamic structure of the interphase nucleus in general, and that of chromosomes and higher-order chromatin in particular, are important elements in the control of gene expression. Here, we investigate the spatial relationship between transcription sites and chromosome territories in the interphase nucleus. Chromosome territories were labeled by FISH, using chromosome-specific DNA whole library probes (Lichter et al. 1988; Jauch et al. 1990). We have analyzed in female primary fibroblasts the localization of transcription sites in relation to the territories of chromosomes X and 19. In addition, we have examined the spatial relationship between total chromatin in nuclei of HeLa cells that express GFP-tagged histone H2B (Kanda et al. 1998) and transcription sites. Sites of transcription were visualized by fluorescent labeling of nascent RNA (Wansink et al. 1993; Jackson et al. 1993).
Our results show that transcription sites are present throughout chromosome 19 (a gene-rich chromosome) and, as expected, in only one of the two X chromosome territories (most likely the Xa). The other X chromosome is devoid of transcription sites (most likely the Xi) and acts as an internal control. FISH labeling of chromosomes reveals a distinct substructure of interphase chromosome territories. Territories consist of areas that are intensely labeled, probably reflecting compact chromatin, and domains that are unlabeled, containing little or no DNA. Analysis of HeLa cells that express GFP–H2B supports this conclusion. Although no individual chromosomes can be distinguished, a similar pattern of intensely labeled domains and areas with little or no GFP signal is observed. These results show that chromosome territories are relatively open structures, rather than compact lumps of chromatin. Strikingly, newly synthesized and nascent RNA accumulates specifically in the interchromatin domains, indicating a distinct compartmentalization of transcriptionally active and inactive chromatin within territories. Each of these conclusions is discussed in detail below.
Structural Preservation during In Situ Hybridization
The relatively harsh FISH procedure may alter the structure of the nucleus and of chromosomes, even after formaldehyde fixation. Therefore, we have thoroughly analyzed the degree of structural preservation during the chromosome painting procedure. Recently, the spatial distribution of acetylated histones in interphase nuclei has been used as a sensitive marker to visualize possible alterations in chromatin structures that are induced by FISH labeling at the light microscopy resolution level (Hendzel and Bazett-Jones 1997). We show that our FISH procedure does not alter the spatial distribution of acetylated histone H4 in fibroblast nuclei. The same is true for the distribution of DNA, stained with Sytox green. These results indicate that the FISH protocol has no major effect on the structure of interphase chromosome territories at the light microscopy resolution level.
This is in agreement with observations of Robinett et al. 1996, who demonstrated that the interphase chromosome structure is well-preserved after in situ hybridization in formaldehyde-fixed cells. In further support of this conclusion, we found that the chromosome painting procedure does not affect the spatial distribution of transcription sites. In addition, the 3-D distributions of centromeres and of PML bodies, compared in one and the same nucleus before and after carrying out the FISH protocol, was hardly affected by the FISH procedure. This is in agreement with other studies demonstrating that the distribution of other subnuclear structures, i.e., nuclear speckles and kinetochores, are not affected by the FISH procedure (Zirbel et al. 1993; Kurz et al. 1996; Hendzel and Bazett-Jones 1997). Finally, in cells expressing histone H2B–GFP, we observe that chromatin is organized in compact domains surrounded by nonchromatin material, very similar to the substructure of chromosome territories observed after FISH labeling. We conclude that the chromosome painting procedure does not significantly alter the structure of the nucleus and of interphase chromosomes at the light microscopical resolution level.
Chromosomal Subdomains
Our results show that chromosome territories have a well-defined substructure and consist of distinct subchromosomal chromatin domains. In optical sections, chromosomes appear as clusters of such domains with a diameter in the range of 300–450 nm. Domains are sometimes interconnected, forming thread-like, folded structures. Consistently, a similar substructure is observed in nuclei after DNA staining with Sytox green. Similar types of spots and threads have been shown by Brakenhoff et al. 1985 in optical sections of mithramycin-labeled mouse neuroblastoma 2A cell nuclei. Chromosomal substructure is not only found in territories of chromosome 19 and Xa, but also in Xi territories, indicating that the presence of chromosomal subdomains does not depend on transcriptional activity.
It may be argued that the compartmentalization in subchromosomal domains that are intensely labeled and domains that appear to contain little or no DNA is an artifact. It cannot be ruled out that the chromosome-specific whole library probe that we used gives incomplete labeling. However, the probe libraries label metaphase chromosomes homogeneously over their complete length (Carter et al. 1990; Telenius et al. 1992). Alternatively, nonlabeled areas may be rich in repetitive sequences, the labeling of which is suppressed in the FISH procedure. However, dual label in situ hybridization with a fluorescently labeled repetitive Cot-1 DNA probe and the X chromosome-specific probe library shows that highly repetitive sequences do not occur in the less intensely labeled domains in chromosome territories (data not shown). Finally, the presumed chromosomal substructure could be an artifact due to the 3-D image restoration protocol. However, careful comparison of optical sections before and after image restoration shows that essentially all intensity modulations in the restored image are also present in the unrestored optical section, albeit in a more blurred state. If chromatin is visualized in a different manner, i.e., in cells expressing GFP-histone H2B, a very similar distribution of strongly labeled domains and areas containing little or no chromatin is observed, compared with the substructure in chromosome territories after FISH labeling. From this, we conclude that chromosome territories have a distinct substructure and are only partially filled with compact chromatin.
In earlier studies in which chromosome painting is used, territories are described as compact objects without a well-defined substructure (Lichter et al. 1988; Cremer et al. 1993; Zirbel et al. 1993; Eils et al. 1996; Kurz et al. 1996). However, close visual inspection of published images in these publications does show distinct intensity modulations in labeled territories, supporting the idea that chromosome territories do have a substructure. Understanding higher-order levels of chromatin structures above that of nucleosomes and 30-nm wide solenoid fibers is limited (Woodcock and Horowitz 1997; Bustamante et al. 1997). It is evident that additional levels of organization exist between the 30-nm chromatin fiber and the 700-μm wide dense sister chromatids of metaphase chromosomes (Manuelidis and Chen 1990; Manuelidis 1990, Manuelidis 1997; Belmont and Bruce 1994; Woodcock and Horowitz 1997). Higher-order structures, such as 100–130-nm chromonema fibers, have been described by Belmont and Bruce 1994 and Robinett et al. 1996, and 240-nm fibers have been observed by Manuelidis 1990. Recently, Zink et al. 1998 showed that chromosome territories in living cells have a similar substructure. Analysis of cell cycle dynamics of a heterochromatic chromosomal region in vivo reveals that various levels of large-scale chromatin organization can be visualized by light microscopy and EM (Li et al. 1998). EM studies have shown that there are two major, intermingled compartments in the nucleus, one containing compact chromatin, whereas the other is mainly filled with proteins and RNA (Belmont and Bruce 1994; Fakan 1994; Woodcock and Horowitz 1995, Woodcock and Horowitz 1997). Our observations are in line with these studies, showing chromatin domains with comparable dimension. It is tempting to speculate that the tightly packed, irregularly shaped substructures represent the chromosome fiber that is folded in a highly convoluted way inside the territory.
Functional Compartmentalization of Chromosome Territories
We find that with the use of FISH labeling, newly synthesized and nascent RNA accumulates in the interchromatin space inside and around chromosome territories. The same result was obtained using cells expressing GFP-tagged histone H2B. This indicates that the observed distribution is not due to labeling artifacts. It may be argued that antibodies recognizing BrU-containing RNA cannot penetrate into the chromosomal subdomains and, therefore, bind only to peripheral RNA. However, this is unlikely since, for instance, anti-DNA antibodies readily label DNA in interphase nuclei (Hendzel et al. 1998) and antihistone antibodies can be used to visualize chromatin in situ (Turner and Fellows 1989; Turner et al. 1989). Our results indicate that transcription sites are located predominantly at the surface of chromosomal subdomains representing compact chromatin.
These results are in agreement with those of Abranches et al. 1998, who showed that transcription occurs throughout wheat nuclei chromosomes in rapidly dividing root cells. Earlier studies, using tritiated RNA precursors and high resolution autoradiography, have shown that perichromatin fibrils, the ultrastructural in situ form of nascent transcripts, are located in the interchromatin space near the surface of compact chromatin domains (Fakan and Puvion 1980; Fakan 1994; Puvion and Puvion-Dutilleul 1996). This has been confirmed in more recent studies, showing that BrU-immunogold–labeled nascent RNA coincides with perichromatin fibrils (Wansink et al. 1996, CMarco et al. 1999). Nascent or newly synthesized RNA is not found inside compact chromatin domains, confirming that transcriptionally active chromatin is exclusively located at or near the surface of compact chromatin domains. Our interpretation is in line with the model for the haploid interphase chromatid structure proposed by Manuelidis et al. (1990), stating that transcriptional active chromatin occurs on locally decondensed chromatin fibers that extend from a compact chromatin fiber.
It has been postulated that the interphase nucleus contains an interchromosomal domain (ICD) space, i.e., the space between the chromosome territories (Cremer et al. 1993; Zirbel et al. 1993). The ICD space is thought to be an interconnected reticular nuclear compartment in which transcription, RNA processing, and RNA transport take place. Our results extend this model by showing that the interchromosomal compartment between chromosome territories is continuous with the interchromatin space inside the territories. In fact, there is no sharp distinction between the surface of a territory and that of compact subchromosomal domains. This reconciles the at-first-sight conflicting results of Kurz et al. 1996 and our observations. They showed that coding DNA is found preferentially at the periphery of chromosome territories, whereas a noncoding genomic locus was found predominantly in the interior of the territory.
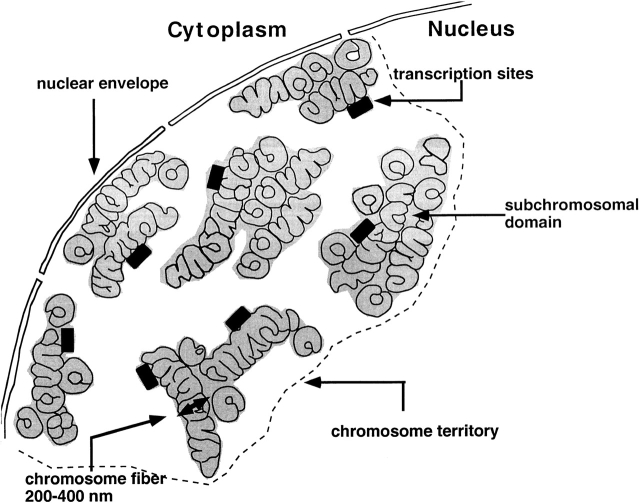
Emerging Model
The following picture is emerging. The chromosome fiber, which may be compacted or locally unfolded to some degree, follows an irregular, convoluted path inside a chromosome territory, very much as suggested by Belmont et al. 1989, Belmont and Bruce 1994, and Robinett et al. 1996. Large-scale chromatin folding is strictly organized in such a way that transcriptionally active DNA is at the surface of the chromosomal fiber (Fig. 6). It is not known whether this compartmentalization of active chromatin is static or dynamic, i.e., whether all actively transcribed genes and those poised for transcription are in the active compartment, or whether a gene, if activated, is moved from the interior of a subchromosomal domain to its surface. Also, whether transcriptionally active loci loop out into the interchromatin space remains to be established. If looping out occurs, it will be only locally and over relatively short distances, since perichromatin fibrils are localized close to the surface of compact chromatin domains. This compartmentalization of active and inactive chromatin allows direct deposition of newly synthesized RNA into the interchromatin space, which contains the molecular machineries for packaging, processing, and transport of RNA. Since the interchromatin space inside territories and the interchromosomal domain between chromosome territories is continuous, such nuclear organization would allow transport of RNA molecules directly to the nuclear pores or to other parts of the nucleus.
Figure 6.
Chromosome territory structure and transcription sites. The cartoon shows a thin section of an interphase nucleus, highlighting a single chromosome territory only. The chromosome fiber is shown, which follows an irregular and convoluted path in the chromosome territory, similar to the chromonema fiber proposed by Belmont and Bruce 1994. In the section, the chromosome fiber is cut perpendicular, oblique, and parallel with respect to the fiber axis. Often, different parts of the chromosome fiber come close together, forming compact subchromosomal domains (gray areas), in which the individual fiber cannot be distinguished. Transcriptionally active chromatin is markedly compartmentalized. Active loci (indicated by black rectangles) are located predominantly at or near the surface of compact chromatin domains. The interchromatin space inside a chromosome territory is continuous with the interchromosomal domain.
This model leads to a number of important questions: what is the molecular basis for the strict compartmentalization of active and inactive chromatin; do proteins that are involved in, e.g., epigenetic silencing and activation, induce large scale remodeling of chromatin in terms of this compartmentalization; does constitutive heterochromatin play a structural role in this higher-order chromatin organization; or is transcriptional activity itself responsible for the observed compartmentalization? Presently, we are using EM approaches to obtain insight in this matter.
Acknowledgments
We would like to thank Dr. B. Turner (University of Birmingham, UK) for kindly providing us with the antibodies recognizing acetylated histone H4 and Dr. J.C.A.G. Wiegant (University of Leiden, The Netherlands) for kindly supplying the fluorescently labeled Cot-1 probe. We are very grateful to Dr. G Wahl (The Salk Institute for Biological Studies, La Jolla, California) for kindly providing us with HeLa cells transfected with histone H2B-GFP fusion protein. Furthermore, we thank Prof. Dr. C.J.F. Van Noorden (University of Amsterdam, The Netherlands) for critical reading of the manuscript and Dr. W. De Leeuw, (CWI, Amsterdam, The Netherlands) for technical assistance in image display.
This work was supported by the EU Biomed II program (project number BMH4-CT95-1139).
Footnotes
1.used in this paper: 3-D, three dimensional; BrUTP, 5-bromouridine 5′-triphosphate; FISH, fluorescent in situ hybridization; GFP, green fluorescent protein; Xa, active X chromosome; Xi, inactive X chromosome
References
- Abranches R., Beven A.F., Aragon-Alcaide L., Shaw P.L. Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 1998;143:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S., Bruce K. Visualization of G1 chromosomesa folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S., Braunfield M.B., Sedat J.W., Agard D.A. Large-scale Chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 1989;98:129–143. doi: 10.1007/BF00291049. [DOI] [PubMed] [Google Scholar]
- Brakenhoff G.J., Van der Voort H.T.M., Van Spronsen E.A., Linnemans W.A.M., Nanninga N. Three-dimensional chromatin distribution in neuroblastoma nuclei shown by confocal scanning laser microscopy. Nature. 1985;317:748–749. doi: 10.1038/317748a0. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose A.B., Holmes S.G., Allis C.D., Broach J.R. Transcriptional silencing in yeast is associated with reduced histone acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Brink M., Humbel B.M., De Kloet E.R., Van Driel R. The unliganded glucocorticoid receptor is localized in the nucleus, not in the cytoplasm. Endocrinology. 1992;130:3575–3581. doi: 10.1210/endo.130.6.1597154. [DOI] [PubMed] [Google Scholar]
- Bustamante C., Zuccheri G., Leuba S.H., Yang G., Samori B. Visualization and analysis of chromatin by scanning force microscopy. Methods: a Companion to Methods in Enzymology. 1997;12:73–83. doi: 10.1006/meth.1997.0449. [DOI] [PubMed] [Google Scholar]
- Carter N., Ferguson-Smith M.A., Affara N.A., Briggs H., Ferguson-Smith M.E. Study of X chromosome abnormality in XX males using bivariate flow karyotype analysis and flow sorted dot blots. Cytometry. 1990;11:202–207. doi: 10.1002/cyto.990110123. [DOI] [PubMed] [Google Scholar]
- CMarco D., Verschure P.J., Martin T.E., Dahmus M.E., Krause S., Fu X.-D., Van Driel R., Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after BrUTP-microinjection. Mol. Biol. Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J.M., Bickmore W.A. The distribution of CpG islands in mammalian chromosomes. Nature Genet. 1994;7:376–382. doi: 10.1038/ng0794-376. [DOI] [PubMed] [Google Scholar]
- Cremer T., Lichter P., Borden J., Ward D.C., Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Human Genet. 1988;80:235–246. doi: 10.1007/BF01790091. [DOI] [PubMed] [Google Scholar]
- Cremer T., Kurz A., Zirbel R., Dietzel S., Rinke B., Schrock E., Speicher M.R., Mathieu U., Jauch A., Emmerich P. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp. Quant. Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- De Jong L., Grande M.A., Mattern K.A., Schul W., Van Driel R. Nuclear domains involved in RNA synthesis, RNA processing and replication. Crit. Rev. Eukar. Gene Expr. 1996;6:215–246. doi: 10.1615/critreveukargeneexpr.v6.i2-3.60. [DOI] [PubMed] [Google Scholar]
- Eils R., Dietzel S., Bertin E., Schrock E., Speicher M.R., Ried T., Nicoud M.R., Cremer C., Cremer T. Three-dimensional reconstruction of painted human interphase chromosomesactive and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent RNA transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fakan S., Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int. Rev. Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Gartler S.M., Goldman M.A. Reactivation of inactive X-linked genes. Dev. Genet. 1994;15:504–514. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- Heard E., Clerc P., Avner P. X-chromosome inactivation in mammals. Annu. Rev. Gen. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne A.W., Crane-Robinson C. Histone acetylation and globin gene switching. Nucl. Acids Res. 1992;20:1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Bazett-Jones D.P. Fixation-dependent organization of core histones following DNA fluorescent in situ hybridization. Chromosoma. 1997;106:114–123. doi: 10.1007/s004120050231. [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Kruhlak M.J., Bazett-Jones D.P. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol. Biol. Cell. 1998;9:2491–2507. doi: 10.1091/mbc.9.9.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A. Chromatin domains and nuclear compartmentsestablishing sites of gene expression in eukaryotic nuclei. Mol. Biol. Rep. 1997;24:209–220. doi: 10.1023/a:1006873614521. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Hassan A.B., Errington R.J., Cook P.R. Visualization of focal sites of transcription within human nuclei. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch A., Daumer C., Lichter P., Murken J., Schroederkurth T., Cremer T. Chromosomal in situ suppression hybridization of human chromosomes and autosomes and its use in clinical cytogenetics. Human Genet. 1990;85:145–150. doi: 10.1007/BF00193186. [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. Histone–GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Koken M.H.M., Puvion-Dutilleul F., Guillemin M.C., Viron A., Linares-Cruz G., Stuurman N., De Jong L., Szostecki C., Calvo F., Chomienne C. The T(1517) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz A., Lampel S., Nickolenko J.E., Bradl J., Benner A., Zirbel R.M., Cremer T., Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.C. Structure and function in the nucleus. Science. 1998;280:547–552. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Li G., Sudlow G., Belmont A.S. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining regionprecise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 1998;140:975–989. doi: 10.1083/jcb.140.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Human Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Manders E.M.M., Stap J., Brakenhoff G.J., Van Driel R., Aten J.A. Dynamics of three-dimensional replication patterns during the S-phase, analyzed by double labeling of DNA and confocal microscopy. J. Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Interphase chromosome positions and structure during silencing, transcription and replication. In: Van Driel R., Otte A.P., editors. Nuclear Organization, Chromatin Structure and Gene Expression. Oxford University Press Inc; New York: 1997. pp. 145–168. [Google Scholar]
- Manuelidis L., Chen T.L. A unified model of eukaryotic chromosomes. Cytometry. 1990;18:8–25. doi: 10.1002/cyto.990110104. [DOI] [PubMed] [Google Scholar]
- Misteli T., Spector D.L. The cellular organization of gene expression. Curr. Opin. Cell Biol. 1998;10:232–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Pinkel D., Landegent J., Collins C., Fuscoe J., Segraves R., Lucas J., Gray J. Fluorescence in situ hybridization with human chromosome-specific librariesdetection of trisomy 21 and translocations of chromosome 4. Proc. Natl. Acad. Sci. USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvion E., Puvion-Dutilleul F. Ultrastructure of the nucleus in relation to transcription and splicingroles of perichromatin fibrils and interchromatin granules. Exp. Cell Res. 1996;229:217–225. doi: 10.1006/excr.1996.0363. [DOI] [PubMed] [Google Scholar]
- Rastan S. Chromosome inactivation and the Xist gene. Cur. Opin. Genet. Dev. 1994;4:292–297. doi: 10.1016/s0959-437x(05)80056-5. [DOI] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L. Macromolecular domains within the cell nucleus. Ann. Rev. Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Stuurman N., De Graaf A., Floore A., Josso A., Humbel B., De Jong L., Van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Telenius H., Pemear A.H., Tunnacliffe A., Carter N.T., Behmel A. Cytogenetic analysis by chromosome genes. Chromosomes Cancer. 1992;4:257–263. doi: 10.1002/gcc.2870040311. [DOI] [PubMed] [Google Scholar]
- Ten Kate T., Van Balen R., Smeulders A.W.M., Groen F.C.A., Den Boer G. SCILAIM, a multi-level interactive image processing environment. Pattern Recogn. Lett. 1990;11:429–441. [Google Scholar]
- Turner B.M., Fellows G. Specific antibodies reveal ordered and cell-cycle related use of histone H4 acetylation sites in mammalian cells. Eur. J. Biochem. 1989;79:131–139. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- Turner B.M., O'Neill L.P., Allam I.M. Histone H4 acetylation in human cells. Frequency of acetylation at different sites defined by immunolabeling with site-specific antibodies. FEBS Lett. 1989;253:141–145. doi: 10.1016/0014-5793(89)80947-0. [DOI] [PubMed] [Google Scholar]
- Van Balen R., Ten Kate T., Koelma D., Mosterd B., Smeulders A.W.M. Scil Imagea multi-layered environment for use and development of image processing software. In: Christensen H.I., Crowley J.L., editors. Experimental Environments for Computer Vision and Image Processing. World Scientific Press; Singapore: 1994. pp. 107–126. [Google Scholar]
- Van der Voort H.T.M., Straster K. Restoration of confocal images for quantitative image analysis. Microscopy. 1985;178:165–181. [Google Scholar]
- Van Driel R., Wansink D.G., Van Steensel B., Grande M.A., Schul W., De Jong L. Nuclear domains and the nuclear matrix. Int. Rev. Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- Wansink D.G., Schul W., Van der Kraan I., Van Steensel B., Van Driel R., De Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase-II in domains scattered throughout the nucleus. J. Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink D.G., Motley A.M., Van Driel R., De Jong L. Fluorescent labeling of nascent RNA in the cell nucleus using 5-bromouridine 5′-triphosphate Celis J.E. Cell BiologyA Laboratory Handbook 1994. 368 374 Academic Press, Inc; San Diego: a [Google Scholar]
- Wansink D.G., Van Driel R., De Jong L. Organization of (pre-) mRNA metabolism in the cell nucleus Mol. Biol. Rep. 20 1994. 45 55b [DOI] [PubMed] [Google Scholar]
- Wansink D.G., Sibon O.C.M., Cremers F.F.M., Van Driel R., De Jong L. Ultrastructural localization of active genes in nuclei of A431 cells. J. Cell. Biochem. 1996;62:10–18. doi: 10.1002/(sici)1097-4644(199607)62:1<10::aid-jcb2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Woodcock C.L., Horowitz R.A. Chromatin organization re-viewed. Trends Cell Biol. 1995;5:272–277. doi: 10.1016/s0962-8924(00)89038-8. [DOI] [PubMed] [Google Scholar]
- Woodcock C.L., Horowitz R.A. Electron microscopy of chromatin. Methods. 1997;12:84–95. doi: 10.1006/meth.1997.0450. [DOI] [PubMed] [Google Scholar]
- Zink D., Cremer T., Saffrich R., Rischer R., Trendelenburg M.F., Ansorge W., Stelzer E.H.K. Structure and dynamics of human interphase chromosome territories in vivo. Human Genet. 1998;102:241–251. doi: 10.1007/s004390050686. [DOI] [PubMed] [Google Scholar]
- Zirbel R.M., Mathieu U.R., Kurz A., Cremer T., Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]