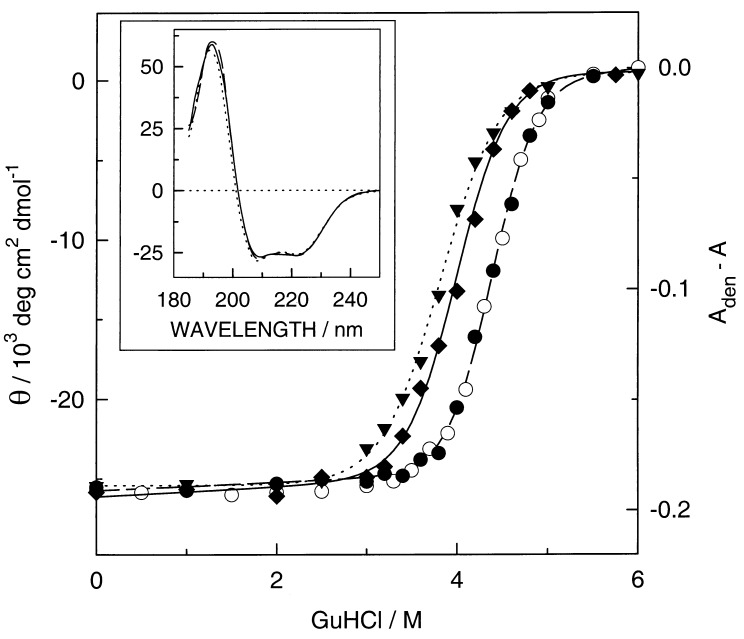
Figure 4.
Guanidine hydrochloride (GuHCl) induced unfolding of the empty MOP2 (▾), heme-Ru-MOP2 (⧫), and heme-MOP2 (•) at 14 μM monitored by CD at 222 nm and of heme-MOP2 at 3.5 μM monitored by the absorbance at 413 nm (○) at 20°C with heme always in the oxidized state. The mean residue molar ellipticity Θ was calculated by dividing the measured ellipticity by the number of 94 residues. The lines are based on the parameters obtained by a nonlinear least-squares fit of the data points to the two-state transition model (33). For the visible region the absorbance difference at 413 nm of the fully denaturated sample minus that of the sample is plotted as a function of the GuHCl concentration. The denaturing experiment was performed as described (19). (Inset) The CD as a function of the wavelength of MOP2 (dotted line), heme-MOP2 (broken line), and heme-Ru-MOP2 (solid line) in 20 mM potassium phosphate, pH 7, at 0 M GuHCl.