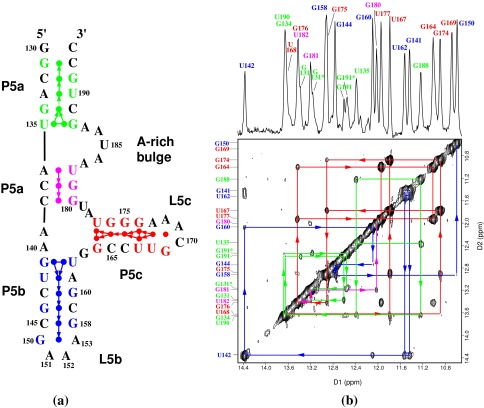
Figure 1.
(a) The secondary structure of the 56-nt P5abc subdomain. The numbering is the same as in the P4–P6 domain. P5abc has 4 bp deleted from the P4–P6 domain between the C145⋅G158 base pair of P5b and the L5b tetraloop GAAA (nucleotides 150–153); the 4-bp deletion causes the discontinuity in the numbering of this region. The disk between each base pair represents the observed imino proton of the base pair. The terminal G130 imino proton resonance is not observed by NMR because of the fraying of the G130⋅C193 base pair. Each arrow represents an NOE connectivity between two imino protons; the direction and color of the arrows match those of NOE walks in the 2D spectrum shown in b. The dotted arrow between the G134 and U190 imino protons indicates our inability to see the NOE between them caused by the near identity of their chemical shifts. (b) The 1D imino proton spectrum (Upper) and 2D 120-ms NOESY spectrum (Lower) of 2.5 mM of P5abc in 10 mM sodium phosphate and 0.01 mM EDTA (pH 6.4) in 90% H2O/10% 2H2O at 10°C. The 1D spectrum was acquired with 4K complex points and processed with a 25°-shifted sine bell squared window function. The 2D data were acquired with 4K points in the D1 dimension and 512 points in the D2 dimension, and were processed with a 40°-shifted sine bell squared function. The arrows connecting diagonal peaks and crosspeaks and the NOE connectivities indicate the spatial arrangement of the imino protons in the secondary structure. The NOE walks are color-coded to the stems in the secondary structure. The 3′ inhomogeneity of the molecule, which is caused by the presence of the N and N+1 species, splits the chemical shifts of imino protons of G131, G191, and U190. Peaks G131* and G191* are from the N+1 impurity molecule, a 57-mer. The peak doubling stops at G134 and does not affect the rest of the molecule.