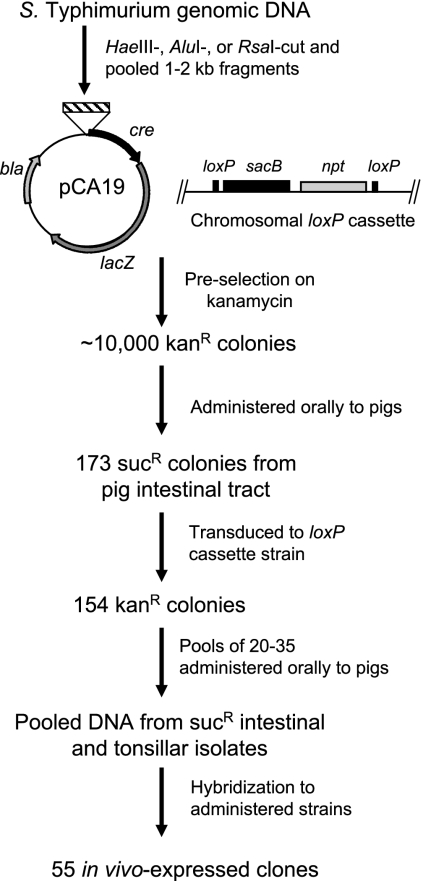
FIG. 1.
Identification of in vivo-induced genes. A genomic library of approximately 10,000 random Salmonella DNA fragments was fused to cre and preselected on kanamycin (Kanr) to eliminate constitutive promoters from the population. The library was then administered to two pigs, and the intestinal contents were cultured on selective medium containing 5% sucrose (Sucr), selecting for bacteria that lost the loxP cassette, along with the intervening sacB, due to the differential expression of cre. Each plasmid was reintroduced into the strain carrying the intact loxP cassette by P22 transduction; of 173 transductants, 154 remained kanamycin resistant. These were divided into groups of 20 to 35 each, and each group was used to infect two pigs. Sucrose-resistant Salmonella isolates were isolated from the ileum and the tonsils, and these were used to make pooled probes by PCR amplifying the cloned fragments of each. Probes were used in colony blots to determine which members of the input pool had reproducibly converted to sucrose resistance after animal infection, producing 55 promoter fragments that were expressed again in both of the pigs that had received the bacterial pool.