Abstract
In this study, we describe the genetic organizations of six and five apparent prophage-like elements present in the genomes of the Lactococcus lactis subsp. cremoris strains MG1363 and SK11, respectively. Phylogenetic investigation as well bioinformatic analyses indicates that all 11 prophages belong to subdivisions of the lactococcal P335 group of temperate bacteriophages.
Prophages are temperate bacteriophages that are inserted into the chromosome of their bacterial host. Whole-genome sequencing projects have revealed that prophage sequences are widespread among bacterial genomes (2, 3, 4, 8, 29, 41-45). Prophages are not only important genetic elements that impart bacterial genome variability. They may confer a diverse array of phenotypic traits to their hosts (sometimes referred to as lysogenic conversion factors), including those that govern the course and pathobiology of bacterial infections (4, 6, 8). Interest in lactococcal phages originally arose from the economic impact of their attacks on Lactococcus lactis cultures that are used for the manufacture of fermented dairy products. The L. lactis species is classified into two subspecies, i.e., Lactococcus lactis subsp. lactis (e.g., L. lactis subsp. lactis IL-1403) and Lactococcus lactis subsp. cremoris (e.g., L. lactis subsp. cremoris MG1363 and L. lactis subsp. cremoris SK11) (34).
Many lactococcal phages have been isolated and have been divided into a distinct number of species (12, 17, 21), as well as subspecies (e.g., P335 [5, 31], c2, and 936 [14, 28]), that all belong to the Caudovirales order (1). Lactococcal phages thus represent a valid model to study genetic organization, population genetics, and mode of evolution. Sequencing of L. lactis subsp. cremoris MG1363 and L. lactis subsp. cremoris SK11 allowed the identification and subsequent analysis of prophage sequences with respect to gene content, transcription profile (in the case of the MG1363 prophages), and comparative genome organization.
Prophage contents of the L. lactis subsp. cremoris MG1363 and SK11 genomes.
As described in previous reports (8, 43), integrases and/or cI repressors are useful markers for the identification of mobile DNA elements such as prophages in bacterial genomes. Recently, the entire sequence of the L. lactis subsp. cremoris MG1363 genome was determined (47), making it possible to characterize the prophages it may contain. Surprisingly, despite the fact that, during its construction, L. lactis strain MG1363 was subject to a prophage-curing scheme (19), six prophage sequences were identified on the basis of significant homology between identified open reading frames (ORFs) and known phage genes. These presumed prophages appear to have integrated into noncoding regions and encompass DNA regions with lengths of 19,053 bp (MG-1), 6,019 bp (MG-2), 42,085 bp (t712), 44,200 bp (MG-3), 18,029 bp (MG-4), and 10,598 bp (MG-5) (names for the putative phages are given in parentheses).
Based on genome length only prophages t712 and MG-3 appear to represent complete phages, whereas MG-1, MG-2, MG-4, and MG-5 are presumed to be incomplete phage elements. In the case of prophage t712 it has been demonstrated that the in silico restriction pattern is identical to that of DNA isolated from active phage obtained from a derivative MG1363 strain lysogenized with the 712 phage, suggesting that this is indeed a complete phage (data not shown). However, it must be noted that the prophage does not equip the host with superinfection immunity against t712, as demonstrated by the sensitivity of MG1363 to t712 phage infection using plaque assays, and there is no apparent lytic response to induction with either mitomycin C or UV light (data not shown). Noninducible, but apparently complete prophages have been identified in the genome sequences of other species, e.g., Lactobacillus johnsonii NCC533 (42). Screening for integrase genes and cI repressors in the sequenced L. lactis subsp. cremoris SK11 strain revealed the presence of five apparently complete or partial prophages, represented by prophage-like sequences of 11,208 bp (SK11-1), 36,971 bp (SK11-2), 32,198 bp (SK11-3), 39,595 bp (SK11-4), and 12,712 bp (SK11-5).
The prophage sequences occupy various positions in the genomes of L. lactis subsp. cremoris MG1363 and SK11; these positions are different in the two strains and also different from those of the previously described prophages of L. lactis IL-1403 (see Fig. S1 in the supplemental material). These differences represent a large portion of the observed genomic differences between L. lactis MG1363, L. lactis SK11, and L. lactis IL-1403 (see Fig. S1 in the supplemental material). When the genomic inversion in L. lactis MG1363 is taken into account (46), the prophages t712, MG3, and MG4 are integrated in the same positions on the genome as the prophages SK3, SK4, and SK5 in the genome of L. lactis SK11. Notably, the putative attachment sites of prophages t712 and SK11-3 are identical, as are those of MG-3 and SK11-4 (see Table S1 in the supplemental material). However, based on similarities they still represent different prophages (see below).
Genome analysis of prophage t712.
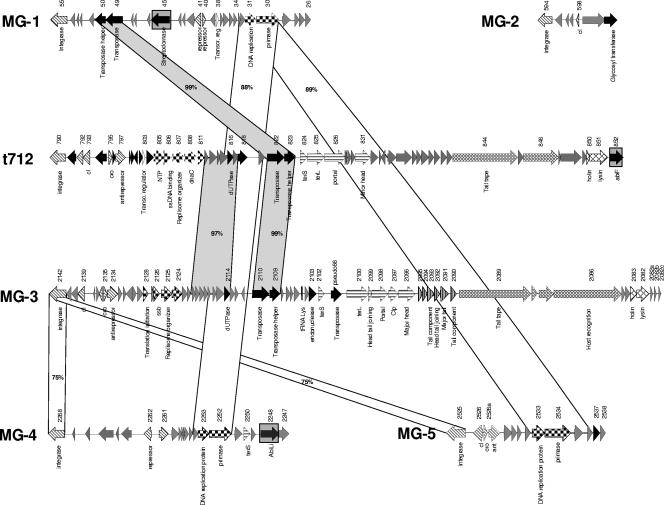
Prophage t712 in L. lactis subsp. cremoris MG1363 extends from ORF 790 (integrase gene) to ORF 852 (abortive infection system-encoding gene) (Fig. 1). These ORFs are flanked by a 13-bp repeat (see Table S1 in the supplemental material), indicative of attL and attR sites. Database matches allowed a subdivision of the t712 prophage genome into functional modules (Fig. 1). The lysogeny module extends from ORF 790 to ORF 797. It includes the integrase gene (ORF 790; 35% identity to an integrase gene of Streptococcus pneumoniae phage MM1), a metalloproteinase gene (ORF 792; 39% identity to a metalloproteinase-encoding gene of Streptococcus pyogenes phage 315.5), a cI-like repressor gene (ORF 793), a cro-like gene (ORF 795), and the putative antirepressor gene ant (ORF 797). Remarkably, the cI- and cro-like genes are not located directly next to each other, as described for many lactococcal phage switch regions (22, 23, 27). Several small ORFs were identified downstream of the lysogeny module, one of which resembles a putative transcriptional regulator-encoding gene (ORF 803).
FIG. 1.
Comparative genome maps of the L. lactis MG1363 prophages MG-1, MG-2, MG-3, MG-4, MG-5, and t712. Similar genes are linked by shading, with the amino acid identities given in percentages. Predicted functions of encoded proteins identified are indicated. The modular structures of the genomes are indicated by different patterns, which indicate their predicted functions: diagonal stripes, lysogeny; large checkerboard, DNA replication; white, transcriptional regulation; horizontal stripes, DNA packaging and head, small checkerboard, tail; narrow vertical stripes, tail fiber; diamonds, lysis modules; grey, similar to bacterial protein; black, hypothetical protein; and wide vertical stripes, tRNA genes. The presumptive lysogenic-conversion genes are highlighted in gray.
The DNA region following the lysogeny module (and extending from ORF 805 to ORF 811) is predicted to encompass the replication module, as it specifies proteins similar to DNA replication proteins from lactococcal and streptococcal phages. Database matches include a gene predicted to encode a nucleoside triphosphate (NTP) binding protein (ORF 805; 62% identity with the NTP binding protein of S. thermophilus Sfi19 phage), a gene that encodes a putative single-stranded-DNA binding protein (ORF 806), a replisome organizer-encoding gene (ORF 808; product has 98% identity with the DNA replication protein of L. lactis bIL 309 phage), and a gene (ORF 811) whose product contains a RusA motif, which has been implicated in DNA replication (35).
A region, containing several anonymous ORFs and several ORFs with a predicted function (ORF 816, ORF 818, ORF 822, and ORF 823) is located between the DNA replication module and the structural protein module of t712. Of note, the ORF 816 product is similar to a deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase), encoded by a wide variety of viral bacterial and eukaryotic genomes. Putative dUTPase genes are without exception found in the genomes of P335-type lactococcal phages (at least the ones that appear to be complete). The dUTPase enzyme catalyzes the cleavage of dUTP into dUMP and pyrophosphate, thereby preventing uracil incorporation into DNA by efficiently decreasing the dUTP/dTTP ratio during phage genome replication (9). The ORF 818-encoded protein is similar to the methyltransferase subunit of a predicted type I restriction modification system found in L. lactis SK11. Finally, the proteins encoded by ORF 822 and ORF 823 resemble an IS21 family of transposase and transposase helper proteins of Enterococcus faecium, respectively. Across the DNA packaging and structural modules, prophage t712 exhibits sequence similarity to sections of corresponding modules found in phages that infect Lactobacillus species. Based on synteny and database matches we identified likely DNA packaging and head morphogenesis genes, possible head-to-tail-joining genes (related to those identified on the L. johnsonii prophage Lj965 genome (39), and putative tail and tail fiber genes. Deduced products of the adjacent lysis cassette genes (ORF 850 and ORF 851) display sequence identity with the putative holin and lysin from L. lactis phage ul36 (23). An ORF (ORF 852) encoding a putative phage resistance-conferring abortive-infection (Abi) protein is located between the lysin gene and the attR site. The ORF 852 protein product contains a typical AbiF motif (COG4823) and displays strong similarity (42% identity) to an abi gene product of S. pyogenes MGAS 10394 (3). The original AbiF-encoding gene is located on lactococcal plasmid pNP40 and has been shown to delay the DNA replication of 936 lactococcal phages (18). Thus, this may represent the first description of a gene encoding an Abi phage resistance system carried by a phage.
Genome analysis of the MG-3 prophage.
The predicted prophage MG-3 of L. lactis MG1363 extends from ORF 2142 (integrase gene) to ORF 2082 (lysin gene) (Fig. 2). These ORFs are flanked by a 23-bp repeat (see Table S1 in the supplemental material), indicating the existence of attL and attR sites. On the basis of identified similarities to other bacteriophages the MG-3 prophage genome can be subdivided into functional modules (Fig. 1). The lysogenic module in this prophage-like element is limited to a region spanning ORF 2142 to ORF 2134. Proteins specified by this region show significant BLAST matches to a phage integrase (the product of ORF 2142; 47% identity to an integrase protein of the L. lactis biL310 phage), a cI-like repressor (encoded by ORF 2139), a Cro-like gene product (ORF 2135), and an antirepressor protein (ORF 2134). Similar to their counterparts in the t712 prophage the cI- and cro-like genes of MG-3 are not arranged in a typical genetic switch region. The ensuing ORFs (extending from ORF 2128 to ORF 2124) constitute the presumed DNA replication module. This region includes ORFs that encode proteins with significant identity to proteins involved in DNA phage replication (16), i.e., a single strand binding protein and a replisome organizer protein. All of these displayed identity scores above 98% with regards to homologous proteins of L. lactis prophage ul36 (23). Many anonymous ORFs (ORF 2123 to 2104) are located downstream of the putative DNA replication module. Interestingly, located among these ORFs is a gene (ORF 2114) whose product has 98% identity with a putative dUTPase of L. lactis ul36 prophage. This gene is very similar to ORF 816 of the t712 prophage. Moreover, comparison of the MG-3 and t712 prophage genome sequences showed significant sequence similarity (above 99%) at the DNA level in the region encompassing ORF 2110 and ORF 2109 of MG-3 and ORF 822 and ORF 823 of t712, resembling a transposase gene and a transposase helper gene, respectively, of E. faecium. Furthermore, a tRNA gene carrying an anticodon (TTT) specific for a Lys residue was found in MG-3 (Fig. 2). Interestingly, tRNA genes are not commonly found in lactococcal phage sequences, and the presence of this tRNA gene in the MG-3 prophage gene may increase the translational efficiency of certain proteins specified by the MG-3 phage or its bacterial host. ORF 2103 is located immediately downstream of the tRNA gene and is predicted to encode an HNH endonuclease (PF01844), homologs of which have been associated with introns in phages (15).
FIG. 2.
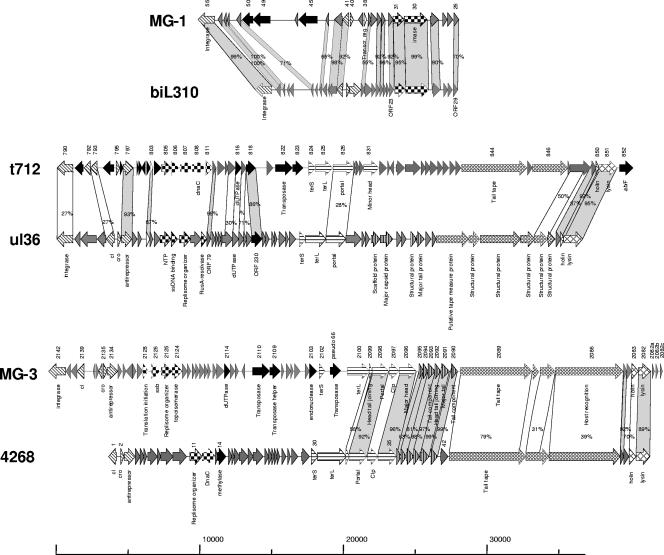
Alignment of the L. lactis MG1363 prophages MG-1, MG-2, MG-3, MG-4, MG-5, and t712 prophages with other lactococcal phages. Representation is as in Fig. 1.
Identification of putative structural modules, such as the head morphogenesis genes (from ORF 2102 to ORF 2096) and the tail morphogenesis genes (from ORF 2085 to ORF 2090), was possible on the basis of the observed homology of gene products to those in other lactococcal phages, i.e., L. lactis biL286 or 4268, and streptococcal phages, i.e., S. thermophilus Sfi19 phage (25, 26). A pseudogene (ORF pseudo66) that is identical to a transposase gene of L. lactis IL-1403 is located between the putative terS and terL genes. The right part of prophage MG-3 (from ORF 2099 to ORF 2082) displays the highest overall similarity (at the amino acid level of the encoded proteins) to the regions of the L. lactis prophage 4268 carrying head morphogenesis and tail morphogenesis gene clusters (37). The general organization of this MG-3 genome region shows a one-to-one correspondence to similarly sized, homologous genes in the 4268 prophage (Fig. 2). The ORF 2089 product contains some conserved elements of a transglycosylase family, which consists of cell wall-hydrolyzing enzymes, including the phage T4 lysozymes and phage-encoded transglycosylases. As ORF 2089 is part of the gene cluster that is predicted to specify tail proteins, the ORF 2089 product may be involved in hydrolyzing the cell wall as part of the initial stages of the phage infection process. The deduced protein of ORF 2086 contains seven collagen-like repeats (Gly-X-Y) and displays similarity to putative host specificity proteins of lactococcal prophage 4268. Collagen-like repeats are motifs with a glycine recurring every third residue that forms a triple helical structure (13). As these motifs were identified in structural proteins that encode the phage host specificity of some phages, we propose that the protein encoded by ORF 2086 encodes the host specificity protein of MG-3.
Immediately downstream of the tail morphogenesis module, a classical host lysis cassette, which includes the presumed holin gene (ORF 2083) and the lysin gene (ORF 2082), was identified, the products of which are similar to those of L. lactis phage 4268 (Fig. 2). Finally, a small set of ORFs (ORF 2082a, ORF 2082b, and ORF 2082c) encoding proteins with similarity to anonymous proteins specified by the L. lactis prophage biL286 and Lc3 are located between the lysin gene and the attR sites.
Genome analysis of the MG-1, MG-2, MG-4, and MG-5 prophage remnants.
The four MG1363 prophages MG-1, MG-2, MG-4, and MG-5 differ from t712 and MG-2, as well as from other complete bacteriophages infecting lactic acid bacteria, by their short genomes and the conspicuous absence of genes required for phage morphogenesis and lysis of the bacterial host. Such genetic functions are also absent in the Escherichia coli satellite phage P4, which depends on helper bacteriophages for the provision of morphogenetic and lytic functions in order to successfully accomplish lytic development (24). Possibly, the four small MG1363 prophages represent satellite phages that rely on “complete” or so-called helper phages for multiplication. This situation may be similar to that suggested for L. lactis IL-1403, in which three small phage remnants, i.e., biL310, biL311, and biL312, whose propagation was proposed to rely on helper phages, are present (10). These small phages may therefore represent cryptic remnants of an as yet uncharacterized phage group.
MG-1 possesses the largest genome (19,053 bp) of this group of presumed prophage remnants, displaying considerable homology and synteny to the L. lactis prophage biL310 genome sequence (Fig. 2). The MG-1 prophage is inserted between a gene encoding a putative mannitol-1-phosphate 5-dehydrogenase and a gene encoding a protein of unknown function. Both ORFs are flanked by a 25-bp repeat (Table 1) that is a likely candidate for the attL and attR sites. The MG-1 prophage genome contains an ORF (ORF 55) which is nearly identical to the integrase gene of L. lactis bil310 prophage. Many anonymous ORFs are present upstream of the integrase gene, in addition to a set of genes (ORF 50 and ORF 49) whose deduced protein products are nearly identical to the predicted transposase helper and transposase encoded by MG-3 and t712. Furthermore, ORF 45 encodes a product that exhibits a 39% amino acid identity (e value of 10−29) to a streptococcal DNase, also called streptodornase, of S. pyogenes MGAS10394 (4).
ORF 41 and 40 of MG-1 show significant database matches with the genes constituting the genetic switch region of the lactococcal phage r1t, while the protein product of ORF 38 is 40% identical to the transcriptional regulator of Streptococcus pneumoniae phage EJ-1. Moreover, ORF 30 and ORF 31 represent elements of the DNA replication module, encoding proteins which display significant similarities to the DNA replication protein and the DNA primase of Enterococcus faecium prophage Lp4 and the lactococcal prophage biL310, respectively.
MG-2 has the shortest sequence of all lactococcal phages analyzed so far. It consists of five phage-related genes, including the integrase gene (ORF 594) and a gene for a cI repressor (ORF 598). Their predicted protein products show 30% and 62% identity, respectively, to the homologous proteins in the L. lactis subsp. lactis IL-1403 prophage biL286. This prophage remnant is flanked by a 46-bp repeat (see Table S1 in the supplemental material) presumed to represent the attL and attR sites. Phage integration complements the 3′ end of a tRNAArg gene carrying a CCG anticodon. When the deduced protein products of the genome of MG-2 were compared to those encoded by other L. lactis phages, the highest similarities were observed with proteins encoded by L. lactis prophage biL286 (see above and data not shown).
The MG-4 prophage remnant is inserted between a putative S-adenosylmethionine-dependent RNA methyltransferase gene and a gene that is homologous to a gene regulating the production of exoamylase and other exoenzymes (46). The prophage sequence is flanked on each side by two 16-bp repeats, indicating the existence of putative attL and attR sites. The predicted integrase gene (ORF 2268) of MG-4 is preceded by five anonymous genes, a gene (ORF 2262) whose product contains a helix-turn-helix motif, and by a gene (ORF 2261) that is similar to a cro-like gene of the L. plantarum prophage Lp4 (42). The last two genes are likely to constitute the genetic switch region of MG-4. Downstream of this presumptive lysogeny module are two genes (ORF 2253 and ORF 2252), whose predicted protein products are similar to a DNA replication protein and to a primase, respectively, of E. faecium prophage Lp4. The MG-4 prophage structural module appears to be limited to ORF 2250, whose protein product is similar to the small terminase protein of L. lactis subsp. lactis prophage biL310. The MG-4 prophage contains a gene (ORF 2248) whose protein product is similar to a protein (AbiLi) that forms part of a two-component abortive-infection system found in L. lactis and Staphylococcus epidermidis ATCC 12228. Although the precise mechanism of AbiLi is unknown, it was shown to prevent 936 phage replication (12).
The MG-5 prophage remnant is located between a gene (ORF 2524) encoding an uncharacterized protein containing a Zn ribbon domain, possibly involved in phosphonate metabolism, and a gene (ORF 2539) encoding a glyceraldehyde-3-phosphate dehydrogenase, while being flanked on either side by a 14-bp repeat (see Table S1 in the supplemental material), thus representing candidates for the attL and attR sites. The presumed MG-5 prophage lysogeny module contains four genes, whose deduced protein products display a high level of protein similarity to integrases (ORF 2525), a cI repressor (ORF 2526), a Cro repressor (ORF 2526a), and an antirepressor protein (ORF 2527). The putative cI- and cro-like genes are divergently oriented, and their products share 43% and 31% identity with a transcriptional regulator of the Cro/cI family of S. pyogenes MGAS10394. Upstream of the lysogeny module, ORF 2533 and ORF 2534 were identified as representing part of the DNA replication module. Their predicted protein products match a DNA replication protein (ORF 2533) of E. faecium phage Lp4 and a primase protein of the L. lactis subsp. lactis prophage biL310 (ORF 2534). The putative DNA replication genes are followed by a number of short anonymous genes and ORF 2537, whose predicted protein shows 54% identity with the experimentally determined activator protein of late transcription of L. lactis subsp. lactis phage TP901-1 (7).
Genome analysis of the L. lactis subsp. cremoris SK11-1, SK11-2, SK11-3, SK11-4, and SK11-5 prophages.
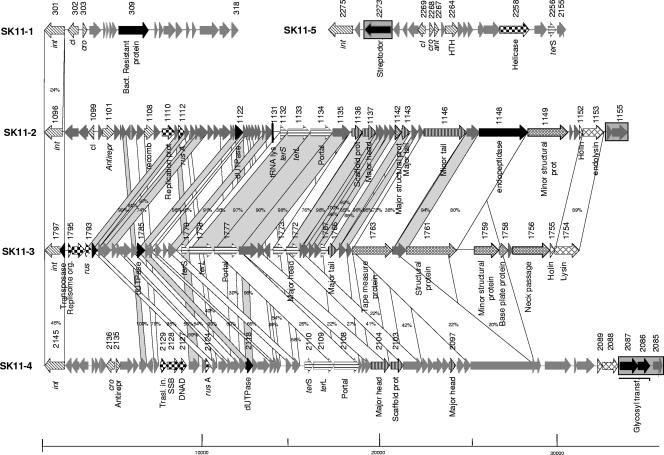
Based on the presumptive prophage genome length and encoded functions only prophages SK11-2, SK11-3, and SK11-4 appear to be complete, while SK11-1 and SK11-5 are more likely to represent prophage remnants. The lengths of all L. lactis subsp. cremoris SK11 prophages was determined indirectly, based on the finding that SK11-1, SK11-2, SK11-3, SK11-4, and SK11-5 prophage sequences are flanked by 24-bp, 13-bp, 14-bp, 23-bp, and 24-bp repeats (see Table S1 in the supplemental material), respectively, in each case suggestive of attL and attR sites.
Genome analysis of the SK11-2 prophage.
Database matches allowed a subdivision of the SK11-2 genome into functional modules typical of lambdoid phages (Fig. 3). The likely extent of the lysogeny module is from ORF 1096 to ORF 1101. This diagnosis was based on clear database matches with integrase and repressor genes constituting the genetic switch region. As in many other temperate phages, the genetic switch region was followed by an antirepressor gene (ORF 1101). Downstream of the lysogeny module, several ORFs (ORFs 1108 to 1112) were predicted to encode proteins with homology to DNA replication proteins, thus defining this genome segment as the likely DNA replication module. A large DNA segment encoding hypothetical proteins with homology to several proteins from P335-type lactococcal phages is located between the DNA replication module and the morphogenesis module. Within this DNA region a gene encoding a putative protein similar to dUTPase of lactococcal Tuc2009 phage was identified. Notably, the following region showed a long non-protein-encoding DNA segment that contained a tRNA gene that recognizes the TTT (Lys) anticodon. Identification of possible structural modules such as head morphogenesis genes (ORF 1132 to ORF 1137), DNA packaging genes (ORF 1138 to ORF 1142), and tail morphogenesis genes (ORF 1143 to ORF 1148) was possible on the basis of observed homology to genes in lactococcal phages (e.g., Tuc2009 and TP901-1 [7, 30]). Separated from the structural modules by two anonymous ORFs, a lysis cassette formed by holin and lysin genes was also identified. Furthermore, two genes, ORF 1154 and ORF 1155, encoding a hypothetical protein and a putative transcriptional regulator protein carrying a helix-turn-helix motif, respectively, were found between the lysis cassette and the putative attR. Notably the putative protein encoded by ORF 1155 shares 56% sequence similarity with a protein encoded by a gene of the biL311 prophage located in a similar genetic constellation.
FIG. 3.
Comparative genome maps of the L. lactis SK11 prophages SK11-1, SK11-2, SK11-3, SK11-4, and SK11-5. Genes sharing similarity are linked by shading. Probable functions of encoded proteins identified by bioinformatics analysis are noted. The degree of amino acid identity is indicated by shading, as described in the legend to Fig. 1.
Genome analysis of the SK11-3 prophage.
The modular organization of the SK11-3 prophage is atypical for temperate phages with low GC content of gram-positive bacteria (Fig. 3). Interestingly, the lysogeny module is limited only to the integrase gene (ORF 1797). It therefore seems that a deletion which removed the genetic switch and flanking regions has occurred in this phage genome. The following ORFs (ORF 1795 to ORF 1793) constitute the putative DNA replication module. Notably, a gene (ORF 1797) which resembles a transposase of Streptococcus thermophilus is located between the lysogeny and the DNA replication modules. Furthermore a gene (ORF 1785) resembling the aforementioned dUTPase gene is present in a genetic constellation typical of P335-type lactococcal phages. Across the DNA-packaging head and tail fiber modules, prophage SK-3 shares sequence similarity with the L. lactis TP901-1 and Tuc2009 phages. A lysis cassette was identified downstream of the tail fiber module, which is constituted by the classical genetic constellation of holin and lysin genes.
Genome analysis of the SK11-4 prophage.
The lysogeny module was predicted to extend from ORF 2145 to ORF 2085 and includes the integrase gene, the genetic switch region, and the antirepressor encoding gene. Notably, the putative cI-and cro-like genes were not located adjacent to each other, as is commonly found for other phages. Downstream of the lysogeny module, many ORFs (from ORF 2134 to ORF 2130) encoded proteins similar to hypothetical proteins of bacteriophage ul36. The downstream ORFs (ORF 2129 to ORF 2127) constitute the likely DNA replication module, which comprises genes encoding a predicted initiation protein, a single-stranded-DNA binding protein, and a replication protein, as based on their displayed similarities to the lactococcal bacteriophage ul36. Several anonymous ORFs (ORFs 2126 to 2111) are placed downstream of the putative DNA replication module. Interestingly, located among these ORFs is a gene (ORF 2118) whose product has 97% identity with the putative dUTPase of the lactococcal ul36 prophage. This gene is very similar to ORF 2114 of the MG-3 prophage. Identification of likely structural modules such as head morphogenesis genes (from ORF 2110 to ORF 2108), DNA packaging genes (from ORF 2104 to ORF 2103), tail morphogenesis genes (ORF 2097 to ORF 2093), and a tail fiber gene (ORF 2090) was possible on the basis of observed homology to genes in Lactobacillus phages. Furthermore, a lysis cassette including the holin- and lysin genes is followed by two ORFs (ORF 2086 and ORF 2087), which encode two putative proteins similar to lactococcal glycosyltransferases. It is possible to speculate that these genes might improve the ecological fitness of the lysogens, thus providing a specific lysogenic-conversion phenotype. One of the possible functions of such glycosyltransferase activity is that they change the sugar composition of a phage receptor (18), which would make this strain insensitive to certain phages.
Genome analysis of the SK11-1 and SK11-5 prophages.
As shown in Fig. 3 the genetic structures of SK11-1 and SK11-5 appear to have been shaped by multiple DNA deletion and rearrangement events. Database matches allowed a functional annotation of only a few ORFs of the SK11-1 prophage genome, comprising the integrase gene (ORF 301) and the genetic switch region divergently transcribed cI- and cro-like repressor genes (ORF 302 and 303). The identical genetic constellation is found in the genome sequences of L. lactis subsp. cremoris MG1363 MG-5 prophage. Notably, another gene (ORF 309), which is similar to a gene encoding a putative bacteriophage resistance protein from S. pyogenes MGAS10270 as well as the L. lactis NCK202.50 strain (20), was identified in the SK11-1 prophage genome.
The prophage-like element SK11-5 consists of a lysogeny module, which includes a possible genetic switch region (ORFs 2268 and 2269), an antirepressor (ORF 2267), and an integrase gene (ORF 2275). Interestingly, the SK11-5 lysogeny module contains an alien gene whose product shows a 90% amino acid identity (E value of 10−177) to the putative streptodornase encoded by the MG-1 prophage (see above). Furthermore, database matches allowed the distinction of a DNA replication module and a DNA-packaging module, which are limited to ORF 2258 and 2255, respectively. The apparently short sizes of these DNA modules could be the consequence of DNA deletion events that have been started at one end of the prophage DNA and ended within the SK11-5 genome. Such findings have already been described for streptococcal prophage sequences, i.e., the prophage remnant Sfi16 of S. thermophilus (40). Large DNA regions of SK11-5 prophage display sequence similarity to the L. lactis subsp. lactis prophage bIL310.
Phylogenetic analysis of the L. lactis subsp. cremoris MG1363 and L. lactis subsp. cremoris SK11 prophages.
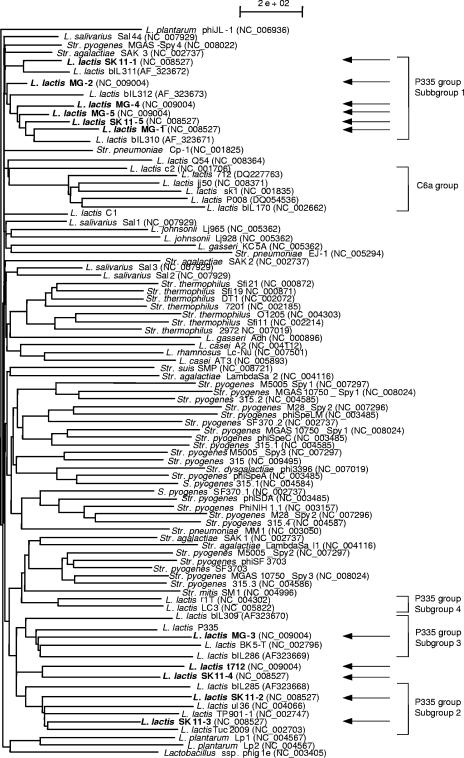
Recently, a sequence-based taxonomic system has been established for inferring phylogeny among phages and prophages through the generation of a proteomic tree (33). We performed such a proteomic tree analysis using the database of phage and prophage sequences with inclusion of the MG-1, MG-2, MG-3, MG-4, MG-5, t712, SK11-1, SK11-2, SK11-3, SK11-4, and SK11-5 sequences. These L. lactis MG1363 and L. lactis SK11 prophage sequences were shown to group with other phage sequences belonging to the lactococcal phage group P335 (Fig. 4).
FIG. 4.
Phage proteomic tree illustrating the relationship between MG-1, MG-2, MG-3, MG-4, MG-5, t712, SK11-1, SK11-2, SK11-3, SK11-4, and SK11-5 prophages and other sequenced phages and prophages.
The proteomic tree produced by this analysis also separates the Lactococcus P335 phage group into four distinct subgroups (Fig. 4). Interestingly, such an analysis showed a clear separation of the cluster formed by r1t and LC3 phages from the other three P335 subgroups (Fig. 4). The proteomic tree produced by the analysis agrees with the classification proposed by Trotter et al. (38) and by Proux et al. (32), who highlighted the close phylogenetic relationship between the Lactococcus and Streptococcus phages.
Comparative lactococcal phage genome analysis.
A comparative genome analysis of the 11 L. lactis prophages described here and the currently available lactococcal phage sequences revealed that they have a very similar genetic organizations, albeit that the genomes are only homologous across less than 30% of their total lengths. The prophage genomes thus appear to be a mosaic of conserved sequences interspersed by nonhomologous regions. Such a genetic arrangement is common among lambdoid phages and reflects combinations of different functional modules, each of which exists in the population as a variable number of disparate versions. Among all P335 phages, the highest resemblance (amino acid sequence and gene order) between the structural proteins of prophage SK11-2 and SK11-3 and the temperate phages Tuc2009 and TP901-1, which form subgroup 2 within the P335 group (Fig. 4). These comparative analyses corroborate the observations on the conservation of the gene order in the morphogenesis/packaging module compared to the variable organization of the early modules (23). These alignments also confirm the extensively discussed high heterogeneity among phages of P335 species genome (7, 10, 23).
Transcriptome analysis of MG-1, MG-2, MG-3, MG-4, MG-5, and t712 prophages.
We assayed L. lactis MG1363 phage gene expression using DNA microarray technology (see the supplemental data). From these results it is obvious that large regions of these prophage genomes are transcriptionally silent during early logarithmic growth of L. lactis MG1363. Regions encoding immunity functions, as well as those encoding lysogenic conversion functions, i.e., putative abi determinants and putative mitogenic factors, were shown to be expressed during logarithmic growth (see the supplemental material).
Conclusions.
The genome sequences of t712, SK11-2, SK11-3, and SK11-4 prophages, with respect to the overall genome organization, were shown to belong to the group of Sfi11-like pac site Siphoviridae (32), whereas the MG-3 prophage belonged to the Sfi21-like cos site Siphoviridae (37). Phylogenetic analysis based on a proteomic tree as well as comparative genome analyses revealed that the MG-1, MG-2, MG-3, MG-4, MG-5, t712, SK11-1, SK11-2, SK11-3, SK11-4, and SK11-5 prophage sequences described here are phylogenetically related, and all belong to the P335 quasispecies. Notably, the obtained proteomic tree hints to the possibility of a further subdivision of the P335 into subgroups, showing that the P335-like phages are a perfect example of polythetic species. Indeed, previous studies based on DNA-DNA hybridization as well as comparative genome analyses already pointed out that the P355 species is composed of interconnected isolates with shared properties or modules (12).
Previous phage DNA transcription studies of Streptococcus, Lactococcus, and Lactobacillus have shown that large parts of the prophage genome are transcriptionally silent during the lysogenic state, while genes near either attachment site were actively transcribed (40-46). Similar patterns were observed for L. lactis prophages MG-1, MG-3, MG-4, MG-5, and t712. Transcription analysis of L. lactis MG1363 cultures in the early stages of growth revealed the presence of mRNA of several of the presumed phage repressor-encoding genes and the lack of expression of the cro-like genes (see the supplemental material).
The gene located between the the lysin gene and attR (which encompasses the presumed lysogenic-conversion region), abiF, was transcribed in the t712 prophage at both early and late growth phases. A previous study identified an alternative candidate for the lysogenic-conversion region within the lysogeny module (40). This would be consistent with the location of ORF 45 in the lysogeny module of MG-1, which encodes a putative streptodornase protein and which was shown to be transcribed during the lysogenic state. In phages infecting bacterial pathogens, this region carries genes that may provide selective advantage to the lysogens (6). It has been reported that, in many dairy prophages, transcription of genes located in the lysogenic-conversion region is more prominent than that of the phage repressor (40), suggesting a physiological function of these prophage genes in the lysogen. The lack of functional characterization of these putative lysogenic-conversion genes makes it difficult to speculate about the exact nature of their encoded functions and specified phenotypes. However, the predicted function of ORF 852 (an abiF homolog) from t712, located within the lysogenic-conversion region and transcribed during the lysogenic state, is that of a phage defense system that interferes with phage multiplication following phage adsorption and DNA injection (for a review see reference 11). Abi-encoding genes are generally plasmid carried, and they are specifically active against phages (36). It is therefore possible that the t712 phage-encoded Abi determinant is beneficial to L. lactis MG1363 by providing increased resistance against phage infection. Similar to prophage t712, the MG-4 prophage contains a possible Abi-encoding gene, one which resembles that encoding AbiLi. To our knowledge the presence of Abi-like genes in prophage genomes represents a novel finding, and future studies will be carried out in order to experimentally investigate the role played by these Abi-like genes in conferring phage resistance.
Supplementary Material
Acknowledgments
This work was financially supported by Science Foundation Ireland through an Investigator grant to D.V.S., by the Italian Award for Outstanding Young Researcher scheme “Incentivazione alla mobilita' di studiosi stranieri e italiani residenti all'estero” and a Marie Curie Reintegration Grant (MERG-CT-2005-03080) to M.V., and by an IRCSET Embark postdoctoral fellowship scheme 2005 to C.C. Udo Wegmann and Claire Shearman acknowledge funding by a CSG grant from the BBSRC.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ackermann, H. W. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, D. J., S. F. Porcella, K. D. Barbian, S. B. Beres, L. E. Philips, J. M. Voyich, F. R. DeLeo, J. M. Martin, G. A. Somerville, and J. M. Musser. 2004. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190:727-738. [DOI] [PubMed] [Google Scholar]
- 4.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Shlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bissonnette, F., S. Labrie, H. Deveau, M. Lamoureux, and S. Moineau. 2000. Characterization of mesophilic mixed starter cultures used for the manufacture of aged cheddar cheese. J. Dairy Sci. 83:620-627. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 7.Brondsted, L., S. Ostergaard, M. Pedersen, K. Hammer, K., and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 8.Canchaya, C., C. Proux, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, S., B. Segelke, T. Lekin, H. Krupka, U. S. Cho, Y. Kim, M. So, C. Kim, C. M. Naranjo, Y. Rogers, M. S. Park, G. S. Waldo, I. Pashkov, D. Cascio, J. L. Perry, and M. R. Sawaya. 2006. Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism. J. Mol. Biol. 314:503-517. [DOI] [PubMed] [Google Scholar]
- 10.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chopin, M. C., A. Chopin, and E. Bidnenko. 2005. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 8:473-479. [DOI] [PubMed] [Google Scholar]
- 12.Deveau, H., S. J. Labrie, M. C. Chopin, and S. Moineau. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 14.Dupont, K., F. K. Vogensen, and J. Josephsen. 2005. Detection of lactococcal 936-species bacteriophages in whey by magnetic capture hybridization PCR targeting a variable region of receptor binding protein genes. J. Appl. Microbiol. 98:1001-1009. [DOI] [PubMed] [Google Scholar]
- 15.Foley, S., A. Bruttin, and H. Brüssow. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol. 74:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley, S., S. Lucchini, M. C. Zwahlen, and H. Brussow. 1998. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology 250:377-387. [DOI] [PubMed] [Google Scholar]
- 17.Fortier, L. C., A. Bransi, and S. Moineau. 2006. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J. Bacteriol. 188:6101-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garvey, P., G. F. Fitzgerald, and C. Hill. 1995. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1990. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J. Bacteriol. 172:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 22.Kenny, J. K., S. Leach, A. B. de la Hoz, G. Venema, J. Kok, G. F. Fitzgerald, A. Nauta, J. C. Alonso, and D. van Sinderen. 2006. Characterization of the lytic-lysogenic switch of the lactococcal bacteriophage Tuc2009. Virology 347:434-446. [DOI] [PubMed] [Google Scholar]
- 23.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 24.Lindqvist, B. H., G. Deho, and R. Calendar. 1993. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol. Rev. 57:683-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucchini, S., F. Desiere, and H. Brussow. 1999. Comparative genomics of Streptococcus thermophilus phage species support a modular evolution model. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchini, S., F. Desiere, and H. Brussow. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232-243. [DOI] [PubMed] [Google Scholar]
- 27.Mahanivong, C., J. D. Boyce, B. E. Davidson, and A. J. Hillier. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahony, J., H. Deveau, S. McGrath, M. Ventura, C. Canchaya, S. Moineau, G. F. Fitzgerald, and D. van Sinderen. 2006. Sequence and comparative genomic analysis of lactococcal bacteriophages jj50, 712 and P008: evolutionary insights into the 936 phage species. FEMS Microbiol. Lett. 261:253-261. [DOI] [PubMed] [Google Scholar]
- 29.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goldstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sulllivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGrath, S., H. Neve, J. F. M. L. Seegers, R. Eijlander, C. S. Vegge, L. Brøndsted, K. J. Heller, G. F. Fitzgerald, F. K. Vogensen, and D. van Sinderen. 2006. Anatomy of a lactococcal phage tail. J. Bacteriol. 188:3972-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moineau, S., M. Borkaev, B. J. Holler, S. A. Walker, J. K. Kondo, E. R. Vedamuthu, and P. A. Vandenbergh. 1996. Isolation and characterization of lactococcal phages from U.S. buttermilk plants. J. Dairy Sci. 79:2104-2111. [Google Scholar]
- 32.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brussow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleifer, K. H. 1987. Recent changes in the taxonomy of lactic acid bacteria. FEMS Microbiol. Rev. 46:201-203. [Google Scholar]
- 35.Sharples, G. J., E. L. Bolt, and R. G. Lloyd. 2002. RusA proteins from the extreme thermophile Aquifex aeolicus and lactococcal phage r1t resolve Holliday junctions. Mol. Microbiol. 44:549-559. [DOI] [PubMed] [Google Scholar]
- 36.Suzek, B. E., M. D. Ermolaeva, M. Schreiber, and S. L. Salzberg. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123-1130. [DOI] [PubMed] [Google Scholar]
- 37.Tangney, M., and G. F. Fitzgerald. 2002. AbiA, a lactococcal abortive infection mechanism functioning in Streptococcus thermophilus. Appl. Environ. Microbiol. 68:6388-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trotter, M., O. McAuliffe, M. Callanan, R. Edwards, G. F. Fitzgerald, A. Coffey, and R. P. Ross, R. P. 2006. Genome analysis of the obligated lytic bacteriophage 4268 of Lactococcus lactis provides insights into its adaptable nature. Gene 366:189-199. [DOI] [PubMed] [Google Scholar]
- 39.van Hijum, S. A., A. de Jong, G. Buist, J. Kok, and O. P. Kuipers. 2003. UniFrag and GenomePrimer: selection of primers for genome-wide production of unique amplicons. Bioinformatics 19:1580-1582. [DOI] [PubMed] [Google Scholar]
- 40.van Hijum, S. A., J. Garcia, de la Nava, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPrep: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241-244. [PubMed] [Google Scholar]
- 41.Ventura, M., and H. Brussow. 2004. Temporal transcription of the virulent Streptococcus thermophilus bacteriophage Sfi19. Appl. Environ. Microbiol. 70:5041-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura, M., A. Bruttin, C. Canchaya, and H. Brüssow. 2002. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology 302:21-32. [DOI] [PubMed] [Google Scholar]
- 43.Ventura, M., C. Canchaya, and H. Brüssow. 2003. Integration and distribution of Lactobacillus johnsonii prophages. J. Bacteriol. 185:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura, M., C. Canchaya, and H. Brüssow. 2004. The prophages of Lactobacillus johnsonii NCC 533: comparative genomics and transcription analysis. Virology 320:229-242. [DOI] [PubMed] [Google Scholar]
- 45.Ventura, M., C. Canchaya, V. Bernini, E. Altermann, R. Barrangou, S. McGrath, M. J. Claesson, Y. Li, S. Leahy, C. D. Walker, R. Zink, E. Neviani, J. Steele, J. Broadbent, T. R. Klaenhammer, G. F. Fitzgerald, P. W. O'Toole, and D. van Sinderen. 2006. Comparative genomics and transcriptional analysis of prophages identified in the genomes of Lactobacillus gasseri, Lactobacillus salivarius, and Lactobacillus casei. Appl. Environ. Microbiol. 72:3130-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ventura, M., C. Canchaya, M. Kleerebezem, W. M. de Vos, R. Siezen, and H. Brüssow. 2003. The prophage sequences of Lactobacillus plantarum strain WCFS1. Virology 316:245-255. [DOI] [PubMed] [Google Scholar]
- 47.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. Kuipers, D. van Sinderen, and J. Kok. 2007. The complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.