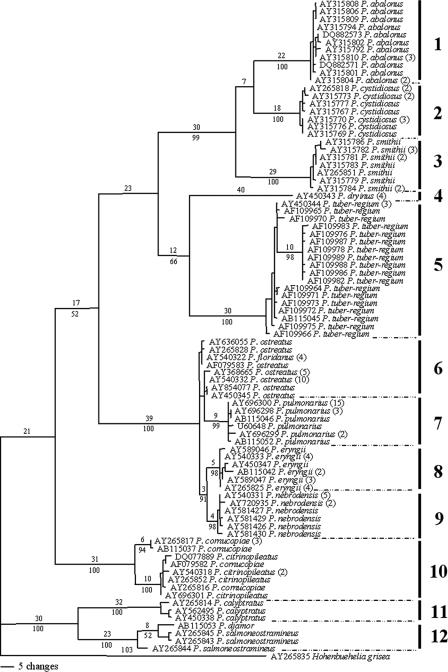
FIG. 2.
One of the 1,288 most parsimonious trees obtained from the analysis of nucleotide sequences of ITS regions (nuclear rDNA). The upper and lower numbers on each branch denote the number of estimated substitutions and the percentage of bootstrap replicates, respectively. Only bootstrap values higher than 50% are shown. Numbers in the brackets after species names are the numbers of sequences that the haplotypes represented. The length of the tree is 701 steps, with a consistency index of 0.6904 and a retention index of 0.9469. Grouping is as follows: group 1, P. abalonus; group 2, P. cystidiosus; group 3, P. smithii; group 4, P. dryinus; group 5, P. tuber-regium; group 6, P. ostreatus; group 7, P. pulmonarius; group 8, P. eryngii; group 9, P. nebrodensis; group 10, P. cornucopiae; group 11, P. calyptratus; and group 12, P. djamor.