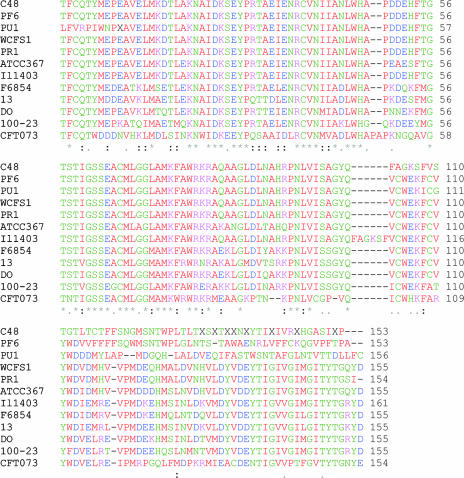
FIG. 2.
Alignment of the internal deduced amino acid sequences of GAD of Lactobacillus paracasei PF6, Lactobacillus delbrueckii subsp. bulgaricus PR1, Lactococcus lactis PU1, and Lactobacillus plantarum C48 with other similar GAD sequences from Lactobacillus plantarum WCFS1 (accession number NP_786643.1), Lactobacillus brevis ATCC 367 (accession number YP_795941.1), Lactococcus lactis subsp. lactis Il1403 (accession number NP_267446.1), Listeria monocytogenes strain F6854 (accession number ZP_00234896.1), Clostridium perfringens strain 13 (accession number NP_562974.1), Enterococcus faecium DO (accession number ZP_00603789.1), Lactobacillus reuteri 100-23 (accession number ZP_01274543.1), and Escherichia coli CFT073 (accession number NP_753818.1). The deduced amino acid sequence was analyzed using ClustalW (1.81).