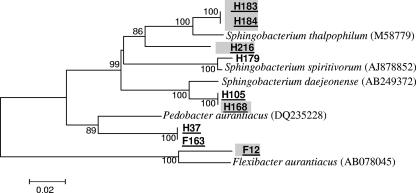
FIG. 2.
Phylogenetic tree of isolates belonging to the Sphingobacteria class. The tree shows the relationship based on partial sequences of the 16S rRNA gene of selected isolates. No special marks on the isolate name indicates no enzymatic activity, gray background indicates lipolytic activity; isolates with names underlined represent new species. The letters F and H in isolate names are the different farms from which the bacteria were obtained. The sequence alignment was performed by means of the CLUSTAL W program, and the tree was generated by the neighbor-joining method with Kimura two-parameter distances in MEGA 3 software. Bootstrap values (from 1,000 replicates) greater than 50% are shown at the branch points. The bar indicates 2% sequence divergence.