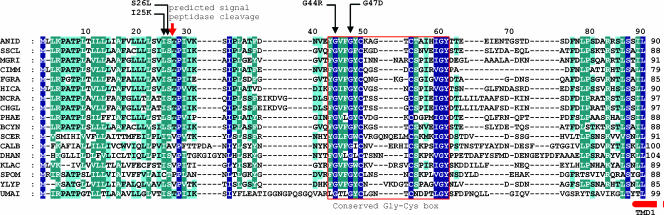
FIG. 1.
The N-terminal region of PalI and Rim9p family members. Multiple sequence alignment of the ∼100 N-terminal residues of PalI/Rim9p homologues in filamentous and yeast ascomycetes and the dimorphic basidiomycete Ustilago maydis. Single-residue substitutions of residues within this region that led to complete or partial loss of function are indicated (Table 2). The position of the signal peptide cleavage site predicted by SignalPeptide 3.0 (http://www.cbs.dtu.dk/services/SignalP/) with a 0.84 probability is indicated by a red arrow. The red box area is the conserved Gly-Cys motif. The start of the first TMD, corresponding to the second hydrophobic region from the N termini of the preproteins (10), is indicated by a broken bar. Conserved residues (according to the Blosum62 matrix) were shaded with navy blue, blue-green, and blue, indicating full, more than 80% but less than full conservation, and more than 60% but less than 80% conservation, respectively. ANID, A. nidulans; SSCL, Sclerotinia sclerotiorum; MGRI, Magnaporthe grisea; CIMM, Coccidioides immitis; FGRA, Fusarium graminearum; HICA, Histoplasma capsulatum; NCRA, Neurospora crassa; CHGL, Chaetomium globosum; PHAE, Phaeosphaeria nodorum; BCYN, Botrytis cinerea; SCER, S. cerevisiae RIM9p; CALB, Candida albicans Rim9; DHAN, Debaryomyces hansenii; KLAC, Kluyveromyces lactis; SPOM, Schizosaccharomyces pombe; YLYP, Yarrowia lipolytica; UMAI, Ustilago maydis.