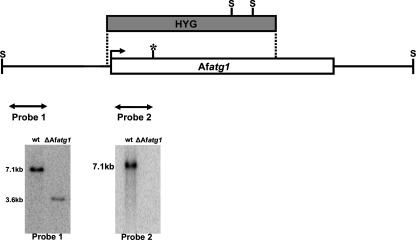
FIG. 1.
Disruption strategy for Afatg1. The split-marker approach was used to create the ΔAfatg1 mutant. Homologous recombination between the disruption cassette and the Afatg1 gene inserted the hygromycin resistance gene (HYG) at the indicate sites (dotted lines). The predicted active site in the kinase domain is shown by an asterisk. Southern blot analysis of SacII (S)-digested genomic DNA by using probe 1 (flanking the disruption cassette) identified the expected 7.1-kb wt band, which was truncated to 3.6 kb in the ΔAfatg1 mutant. A second probe derived from the deleted region (probe 2) confirmed that no duplication had occurred.