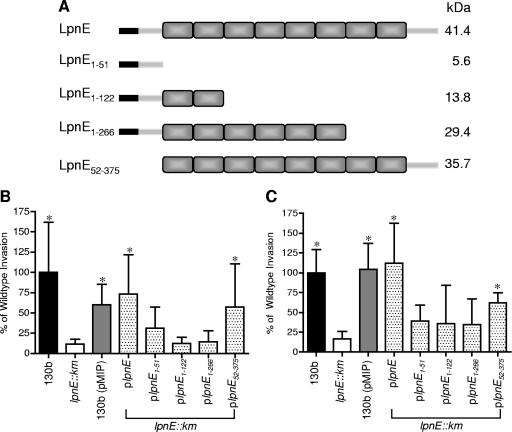
FIG. 4.
(A) Schematic representation of L. pneumophila SLR protein LpnE and truncated variants created for this study. Shaded rectangles represent the SLR regions, and black rectangles signify the predicted N-terminal 22-amino-acid signal peptide. The truncations were used to complement L. pneumophila lpnE::km, and resulting strains were examined for uptake by THP-1 macrophages (B) and A549 epithelial cells (C). Data are expressed as percentages of the amount of inoculum that was intracellular following a 2-h infection and 1-h gentamicin treatment and are means ± standard deviations for at least three independent experiments. *, significantly different from the lpnE::km mutant (P < 0.05; unpaired two-tailed t test).