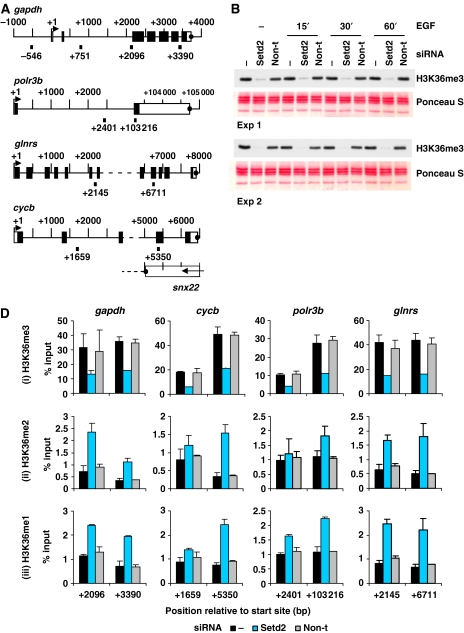
Figure 5.
Setd2 is responsible for H3K36 trimethylation at IE and housekeeping genes. (A) Schematic representation of the gapdh, polr3b, glnrs and cycb housekeeping genes showing regions amplified by primers used for real-time PCR. Primer positions shown indicate 5′ position of the forward primer relative to the transcription start site. Exons are represented by boxes, unfilled for untranslated regions and filled for translated regions. Transcription termination sites are shown as filled circles. (B) Cells were untransfected (−) or transfected with Setd2 or non-targeting (non-t) siRNA. Cells were quiesced 24 h later, and after a further 24 h were unstimulated (−) or stimulated with EGF (50 ng/ml) for 15–60 min. Formaldehyde crosslinked mononucleosomes were prepared and aliquots of each sample were heated to reverse the crosslinks, separated by 15% SDS–PAGE, transferred to PVDF and immunoblotted with an H3K36me3-specific antibody. Membranes were stained with Ponceau S before immunoblotting to verify even loading. (D) Aliquots of formaldehyde crosslinked mononucleosomes from unstimulated cells prepared as in (B) were used in ChIP assays with H3K36me3- (i), H3K36me2- (ii) and H3K36me1- (iii) specific antibodies. Recovery of gapdh, cycb, polr3b and glnrs coding region sequences were quantified by real-time PCR. Average % input recoveries and s.d. from two independent experiments are plotted.