FIG. 2.
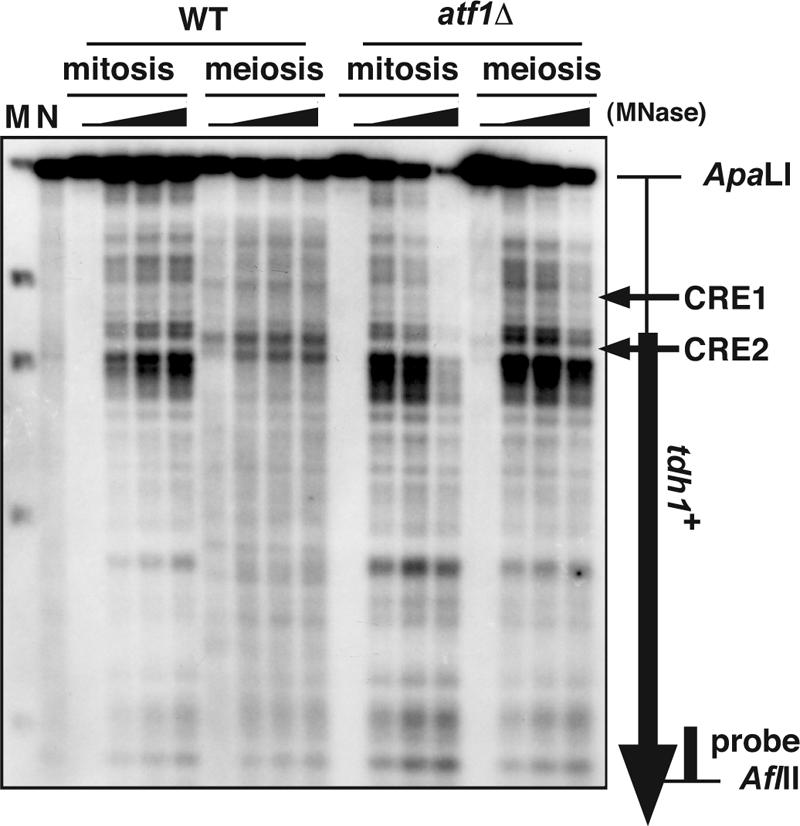
Meiotic chromatin remodeling in the tdh1+ locus depends on Atf1. MNase-digested chromatin DNAs from diploid strains D20 (wild type [WT]) and WSP779 (atf1Δ) were analyzed as in Fig. 1. The atf1Δ samples were slightly overdigested, since the atf1Δ mutant is more sensitive to Zymolyase treatment that allows increased permeation of MNase. The vertical and the horizontal arrows indicate the tdh1+ ORF and the position of the CRE sequences, respectively. Lane M, marker λ, EcoT14I digested (Takara); lane N, MNase-digested naked S. pombe genome DNA.
