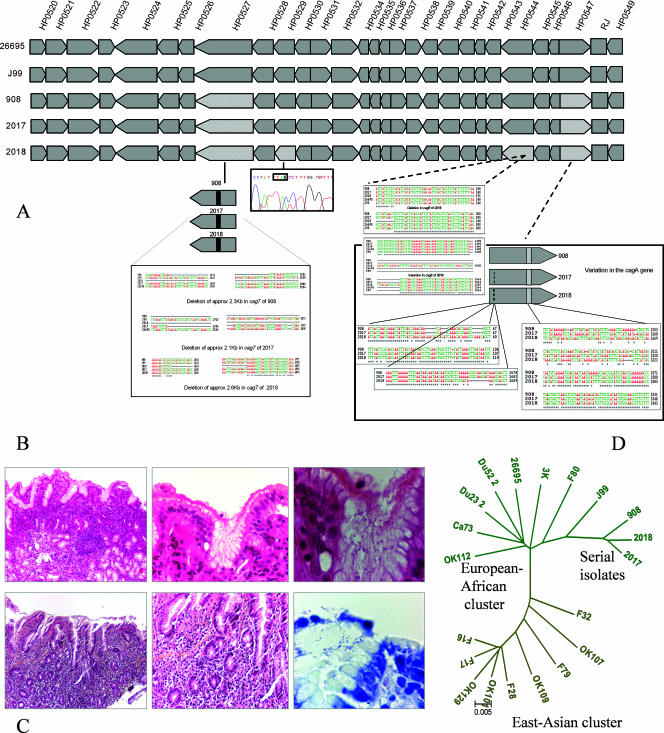
FIG. 1.
(A) Genomic alignments based on pan-island sequencing of cagPAIs from isolates 908, 2017, and 2018 and their comparison with sequences from the two sequenced strains, namely, J99 and 26695. For details of sequencing strategy and alignment methods, see the text. (B) Histopathology of biopsy sections corresponding to isolate 908, originally obtained in 1994 from three antropyloric biopsy samples that revealed duodenal ulcer (inflammatory infiltrate [3+] with polymorphs [3+] and numerous H. pylori on the surface [3+]). (C) Endoscopic diagnosis revealed duodenal ulcer, while histopathological examination of two gastric biopsy samples—one each from antrum (inflammatory infiltrate [2+] with eosinophils, atrophy [+], no metaplasia, no lymphoid follicle, and the presence of H. pylori [2+]) and corpus (slight superficial gastritis)—revealed relatively low-grade pathology in 2003. Isolates 2017 and 2018 corresponding to antrum and corpus, respectively, were successfully cultured from this source. (D) Bifurcating neighbor-joining tree based on whole cagPAI sequence comparison. The clonal association among isolates 908, 2017, and 2018 clearly reveals that they belonged to the same founder strain that shares genogroup affinity with J99 strain (HspSAfrica).