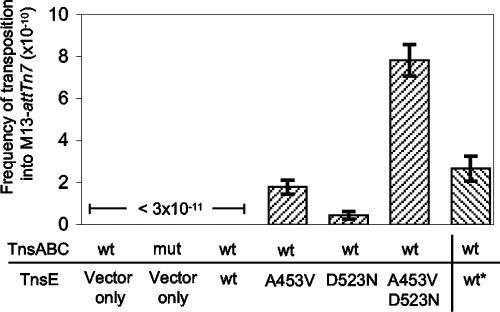
FIG. 1.
Frequency of Tn7 transposition into the M13-attTn7 genome in E. coli in various genetic backgrounds. Wild-type TnsABC was expressed from pCW15 (wt) or a pCW15 derivative with the TnsCA225V mutation (mut) (24). pTA106 was included as a vector control (vector only). Wild-type or mutant derivatives of TnsE, TnsEA453V (A453V), TnsED523N (D523N), or the double mutant TnsEA453V+D523N (A453V D523N) were expressed from pJP103. Wild-type TnsE was expressed with pJP104 (wt*). M13-attTn7 bacteriophage containing mini-Tn7 in strains expressing wild-type TnsABC without TnsE, in the mutant TnsABCA225V, or with TnsABC with wild-type TnsE expressed from pJP103 was never identified (<3 × 10−11). A very low background of spontaneous nalidixic acid-resistant cells (∼10−11) was found in the assay where the F′ Tn10 had mated into residual host strains from the XL1-Blue strain. These were easily identifiable because they contained all of the JF55 chromosomal and plasmid makers (i.e., resistance to rifampin, chloramphenicol, and ampicillin but sensitivity to gentamicin via loss of the pOX-Gen by replicon exclusion). Restriction analysis and DNA sequencing absolutely confirmed that the mini-Tn7 element was not contained on the M13-attTn7 plasmid. Error bars show the standard errors of the means (n = 9).