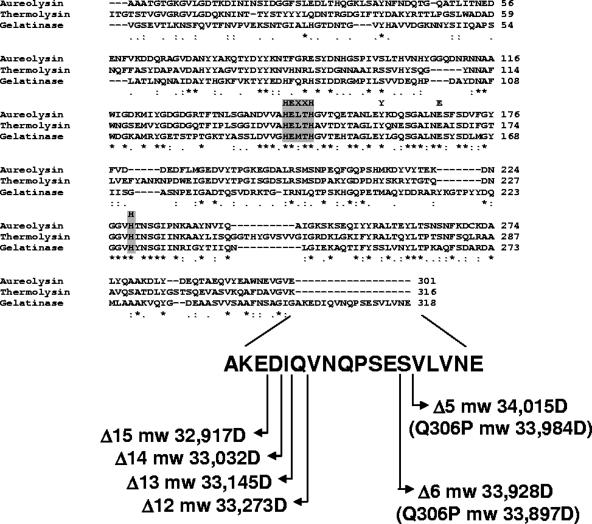
FIG. 1.
Amino acid sequence alignment of members of the M4 family of bacterial metalloproteases. The amino acid sequences of aureolysin of S. aureus, thermolysin of B. thermoproteolyticus, and gelatinase of E. faecalis were aligned using the ClustalW program. The active site residues are indicated by the gray boxes. Symbols: asterisks, identical residues in all sequences; colons, conserved substitutions; periods, semiconserved substitutions. The sequence of the C-terminal 18 amino acids is shown at the bottom along with the calculated molecular masses of GelE mutant proteins lacking the last 5 (Δ5), 6 (Δ6), 12 (Δ12), 13 (Δ13), 14 (Δ14), and 15 (Δ15) amino acids. The predicted molecular masses (mw) were obtained with the ExPASy pI-Mw tool. The predicted molecular masses of the GelE Q306P mutant proteins lacking the last five or six amino acids are indicated in parentheses. D, daltons.