Abstract
Weed plants characteristic for potato and hop fields have not been considered in the past as potential hosts that could transmit and lead to spreading of potato spindle tuber (PSTVd) and hop stunt (HSVd) viroids, respectively. To gain insight into this problem, we biolistically inoculated these weed plants with viroid populations either as RNA or as cDNA. New potential viroid host species, collected in central Europe, were discovered. From 12 weed species characteristic for potato fields, high viroid levels, detectable by molecular hybridization, were maintained after both RNA and DNA transfers in Chamomilla reculita and Anthemis arvensis. Low viroid levels, detectable by reverse transcription-PCR (RT-PCR) only, were maintained after plant inoculations with cDNA in Veronica argensis and Amaranthus retroflexus. In these two species PSTVd concentrations were 105 and 103 times, respectively, lower than in tomato as estimated by real-time PCR. From 14 weeds characteristic for hop fields, high HSVd levels were detected in Galinsoga ciliata after both RNA and DNA transfers. HSVd was found, however, not to be transmissible by seeds of this weed species. Traces of HSVd were detectable by RT-PCR in HSVd-cDNA-inoculated Amaranthus retroflexus. Characteristic monomeric (+)-circular and linear viroid RNAs were present in extracts from weed species propagating viroids to high levels, indicating regular replication, processing, and circularization of viroid RNA in these weed species. Sequence analyses of PSTVd progenies propagated in C. reculita and A. arvensis showed a wide spectrum of variants related to various strains, from mild to lethal variants; the sequence variants isolated from A. retroflexus and V. argensis exhibited similarity or identity to the superlethal AS1 viroid variant. All HSVd clones from G. ciliata corresponded to a HSVdg variant, which is strongly pathogenic for European hops.
Viroids are the smallest known plant pathogens consisting solely of a circular, noncoding RNA ranging from 246 to 463 nucleotides (nt) that incite plant diseases of considerable economic importance (for reviews, see references 2, 3, 6, 10, and 36). There are at least 28 different viroid species classified and listed in biological databases (7, 30). Viroid pathogens are known to be transmissible by mechanical injury of host cells and, therefore, also by the tools or agrotechnique used for agricultural treatments of crops or during picking operations (3). While there is no doubt about the spread of most viroids from plant to plant or from crop to crop as mediated by humans, the existence of wild-plant viroid reservoirs (3) as sources of viroid contaminations could never be shown. Various weed plants characteristic for the particular agrobiotopes and fields could, however, represent a potential danger as reservoirs, “cultivators” or transmitters if they were susceptible to viroid infections. In this respect it is important to note that viroid pathogens form populations of molecular variants that conform to the quasispecies concept (5). Because viroids represent one of the most rapidly evolving biological systems known (4), molecular variants of viroids may serve as a source of adaptations to new hosts, including uncontrolled weed species.
In particular, hop stunt viroid (HSVd) and potato spindle tuber viroid (PSTVd) are two examples of widespread pospiviroids (for viroid classification, see reference 7) forming quasispecies, and some of their sequence variants are strongly pathogenic for cultured plants (for a review, see reference 10). For instance, the grapevine variant of HSVd (HSVdg) was transmitted accidentally to hop causing devastation of hop fields in Japan (see, for example, reference 31). In our previous study (21) we showed that identical HSVdg variants are present in the Czech Republic and Slovenia in local private grapevine gardens in close neighborhood to hop fields. In these gardens the HSVd incidence reaches more that 65% (24). This incidence represents a potential danger provided the existence of susceptible transmitter(s), for example, some common weed plants as viroid “reservoirs.” The experimental host range of PSTVd includes about 160 species, mostly from Solanaceae but also a few species scattered among 10 other families (33; for a review, see reference 2), suggesting also a broad potential of this viroid to spread to various species.
In the present study we tried to inoculate weed plants characteristic for potato and hop fields with complex populations of PSTVd and HSVd, respectively, and to test the potential role of weed plants in transmitting viroid infections to other species. Compared to the standard experimental inoculation method for viroids with Carborundum as an abrasive agent, biolistic inoculation using a gene gun is highly advantageous (25, 26). For example, individual sequences in a viroid population that can be coprecipitated to microprojectiles and delivered at once as a mixture to individual cells have comparative starting conditions for propagation. In addition, using the biolistic approach, one can infect plants or specific tissues that can hardly be inoculated by conventional methods. Indeed, PSTVd and HSVd can propagate in several new experimental hosts, which might form a reservoir for the natural spreading of viroids. Upon the experimental transmission to the new hosts, the viroid sequences underwent sequence changes, which we describe in detail. Some aspects connected to viroid host range and “low-level” viroid populations are discussed.
MATERIALS AND METHODS
Plant species and plant cultivation conditions.
Seeds from the following plant species were collected on potato fields in the surroundings of the city Havlčkºuv Brod (Czech Republic): Amaranthus retroflexus L., Atriplex sagittata Borkh., Chenopodium album L., Anthemis arvensis L., Chamomilla recutita L., Sonchus arvensis L., Galinsoga parviflora Cav., Capsella bursa-pastoris (L.) Med., Stellaria media (L.) Vill., Erodium cicutarium (L.) L'Hér., Plantago major L., and Veronica agrestis L. From hop gardens surrounding the city of Žatec (Czech Republic) we collected seeds from the following major weed species: Amarantus retroflexus L., Atriplex nitens Schkuhr, Artemisia vulgaris L., Cirsium arvense (L.) Scop. Galinsoga ciliata (Raf.) Blake, Senecio vulgaris L., Sonchus arvensis L., Taraxacum officinale Web., Capsella bursa-pastoris (L.) Med., Erysimum cheiranthoides L., Epilobium parviflorum Schreb., Galium aparine L., Urtica urens L., and Urtica dioica L. The following indicator plants were used to analyze viroid infections: Lycopersicon esculentum cv. Rutgers, Nicotiana benthamiana, Humulus lupulus cv. Osvald's 72, Cucumis sativa, and Gynura aurantiaca. H. lupulus, and G. aurantiaca were propagated vegetatively. Plants were maintained in climate boxes at a temperature of 25 ± 3°C under natural light in the period from March 2005 to October 2006 with supplementary illumination (90 μmol m−2 s−1 photosynthetically active radiation) to keep a 16 h-day.
Preparation of viroid inocula and plant inoculation.
For plant inoculation with PSTVd, we used either RNA or cDNA inocula prepared by mixing equimolar quantities of nucleic acids of the following viroid strains: AS1 (GenBank accession code [AC]: AY518939), AS3 (AC: AY673974), RG1 (AC: U23058), KF440-2 (AC: X58388), M3/M1 (AC: AF459007), severe (32), intermediate (AC: V01465), QFA (AC: U23059), and KF5M5 (AC: M93685). This strain list is given in descending order of pathogenic symptoms produced in the indicator plant Lycopersicon esculentum cv. Rutgers (for details, see references 23 and 35). For HSVd inoculation we used a population of HSVdg variants naturally occurring in grapevines in the territory of the Czech Republic as described earlier (24). Inocula were prepared according to methods described previously (25): RNA inocula were prepared by fractionation of 2 M LiCl-soluble nucleic acids with 12 to 20% PEG 6000, and cDNA inocula were prepared as StyI- and EcoRI-cleaved full-length cDNAs of PSTVd and HSVd, respectively. The resulting PSTVd and HSVd populations were immobilized on gold microcarriers (1 μm) using a modified calcium-mediated precipitation protocol (25). The Helios GeneGun system from Bio-Rad was used for biolistic inoculation. Each plant was inoculated twice each time with 2 ng of native viroid RNA or 200 ng of DNA at a pressure of 120 lb/in2 at a distance of approximately 1 cm from the GeneGun spacer. Plants were inoculated on the stage of three true leaves. Attached plant leaves were inoculated while supported with thick cardboard paper. After inoculation, plants were immediately transferred into polyethylene bags to prevent drying of the shot-wound leaf area. Treated plants were further conditioned by shading them for 24 h and afterward by cultivation in holed bags for the next 2 days in the clima boxes under standard conditions as described above. For mechanical inoculation we used 20 μl of inoculum in 0.04 M sodium phosphate buffer (pH 7.6) per leaf and Carborundum 500 mesh (Benátky and Jizerou, Czech Republic) as an abrasive. Unless stated otherwise, eight plants were used for infectivity tests.
Viroid detection procedures and quantification using real-time PCR.
For the reverse transcription-PCR (RT-PCR) and real-time PCR, total RNA was isolated from 100 mg of plant leaf tissue by using CONCERT (plant RNA purification reagent; Invitrogen) following RNA purification and DNA cleavage on columns (RNeasy Plant Total RNA kit; QIAGEN, Germany). RT-PCR amplification for viroid detection and cloning was performed by using Titan One-Tube RT-PCR (Roche) including a high-fidelity Pwo polymerase (Roche Molecular Biochemicals). The primers PSTVds I (5′-aC337CAAGGGCTAAACACCCTCGC-3′) and II (5′-aC343CTTGGAACCGCAGTTGGTTC-3′) were used for full-length PSTVd amplification; the primers HSVde I (5′-aA12GAATTCCCCAGAGGGGCTCA-3′) and II (5′-aG5GAATTCTCGAGTTGCCGC-3′) were used for hop stunt viroid amplification (25). The nonspecific adenine in each primer (indicated by a small letter “a”) was designed to facilitate the cleavage of cDNA fragments; restriction sites encoded in the primers are underlined, and the positions are numbered corresponding to the respective viroid sequence. RT was run for 30 min at 52°C, and after 2 min of denaturation at 94°C the PCR was started with cycles of 30 s at 94°C, 30 s at 58°C, and 60 s at 68°C. RT-PCR was carried out for 38 cycles.
Dot blot hybridizations were performed as described previously (28) with full-length PSTVd or HSVd [32P]dCTP-labeled probes. The lower detection limit of this method is about 0.03 pg per mg of fresh mass (28). For hybridization analysis of PCR products, cDNA fragments were electrophoresed in 2% agarose gels, transblotted to nylon membranes by using alkaline blotting, and hybridized to [α-32P]dCTP-labeled viroid cDNAs as probes.
Circular and linear viroid RNAs were analyzed in 6% polyacrylamide gels containing 30:1 acrylamide-bisacrylamide (wt/wt), 89 mM Tris-borate buffer, 0.24 mM EDTA (1× Tris-borate-EDTA) (pH 8.3), 0.1% TEMED (N,N,N′,N′-tetramethylethylenediamine), 8 M urea, 2% glycerol, and 0.06% ammonium persulfate. Samples were electrophoresed at a constant voltage of 220 V and a temperature of 60°C. Separated RNA was transblotted onto a nylon membrane (charge modified, 0.2-μm pore size; Sigma) using a semidry blotting procedure in Tris-borate buffer and hybridized to [α-32P]dCTP-labeled viroid cDNAs as described earlier (28).
Real-time PCR quantification of PSTVd was performed using the primers RT-R (5′-C189TTTCTTCGGGTGTCCTTCC208-3′) and PCR-R (5′-C289TCGGGAGCTTCAGTTGTTTC269-3′). This PCR led to amplification of a 101-bp product. The 7SL RNA product was used as a constitutive control. Approximately 301 bp of 7SL cDNA was amplified using the primers α (5′-TGTAACCCAAGTGGGGG-3′) and anti-β (5′-GCACCGGCCCGTTATCC-3′) (22). For RT, StrataScript reverse transcriptase 600085 was used according to the manufacturer's recommendations. Reaction mixtures contained 2 μg of total RNA (2 μl), 1 μl of 20 μM RT-R primer, 2 μl of 10× StrataScript buffer, 3 μl of deoxynucleoside triphosphate mix (20 mM), 0.5 μl of RNasin inhibitor (40 U), and 0.5 μl of StrataScript reverse transcriptase (50 U; Stratagene) in 20-μl reactions. After 1 h of incubation (PTC100; MJ Research) at 48°C the real-time PCR mixtures were prepared. This mix contained 10.3 μl of deionized water, 12.5 μl of 2× master mix (brilliant SYBR green QPCR master mix 600548; Stratagene), 0.1 μl of PCR primers (100 mM), and 2.0 μl of the cDNA template in a total volume of 25 μl. This mix was given onto a multiple samples PCR well-plate and reactions were performed in an MX 3005P (Stratagene) using the following protocol: 1 cycle at 95°C for 10 min for polymerase activation, followed by 40 cycles at 95°C for 30 s, 55°C for 45 s, and 72°C for 30 s, with a melting curve program (heating rate of 0.1°C per s and a continuous fluorescence measurement). Viroid detection and quantification was performed 25 days postinoculation.
Other methods.
cDNA probes were labeled with [α-32P]dCTP using the Redivue [α-32P]dCTP (3,000 Ci/mmol) Rediprime II random prime labeling system (Amersham Pharmacia Biotech, Freiburg, Germany). The autoradiograms were scanned by using a Typhoon PhosphoImager (Amersham Biosciences, Sunnyvale, CA) and quantified by using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
For sequencing, cDNA fragments were isolated from the gel by use of the QIAGEN gel extraction kit (QIAGEN, Hilden, Germany) and cloned in vector pCR-Script SK(+) (pCR-Script cloning kit from Stratagene, La Jolla, CA). For each progeny preparation, eight clones were sequenced with an automatic ALF II system (Amersham Pharmacia Biotech, Freiburg, Germany) by using a sequencing kit with Cy5-labeled standard primers. Sequence data analysis was carried out with the computer program DNASIS (version 2.5).
Consensus secondary structures of PSTVd sequences were predicted with the help of a recent version (www.biophys.uni-duesseldorf.de/construct3/) of ConStruct (18). Single structure predictions were performed with RNAfold version 1.6.1 (12) at 37°C. For analysis of a phylogenetic relationship between PSTVd strains used for infection and the resulting progeny, the tool SplitsTree4 was used (13).
RESULTS
Biolistic inoculation of weed species characteristic for potato and hop fields with populations of PSTVd and HSVd.
In order to analyze the ability of PSTVd and HSVd to infect weed plants, we used biolistic transfer of complex viroid populations. Populations of viroid sequences were used because some sequences could have higher adaptability and fitness than others in particular viroid-host combinations and could increase the probability of infection of thus-far-unidentified hosts. The PSTVd population was prepared by mixing equimolar quantities of various viroid strains (see Materials and Methods). As HSVd inoculum, a naturally occurring population isolated from grapevines was used as described earlier (24). Simultaneously, the biolistic approach could eliminate some barriers connected to initial viroid adsorption on cell membranes and transport to nuclei. Based on our previous studies with some Brassica species (25), where only PSTVd transfer as cDNA has been efficient for infection, RNA as well as cDNA inocula were prepared. Although cDNA inoculation does not correspond to a natural viroid infection, our approach increases the chance to identify potential hosts among analyzed weed plants.
Seeds from potato- and hop-field-specific weed species were collected for inoculation experiments (see Materials and Methods). The spectrum of analyzed weeds does not represent all weed plants found on the corresponding agrobiotopes, but only those that were successfully germinated and cultivated under our experimental conditions. This ensemble included 12 and 14 plant species from potato and hop fields, respectively (Tables 1 and 2). Several viroid-susceptible plant species, including tomato, N. benthamiana, hop, cucumber, and G. aurantiaca (Tables 1 and 2), were used as indicators of infection.
TABLE 1.
Detection of PSTVd infection in inoculated weed species from potato fields
| Plant speciesa | Family | PSTVd inoculum and detectionb
|
|||
|---|---|---|---|---|---|
| RNA
|
StyI cDNA fragments
|
||||
| Dot blot | RT-PCR | Dot blot | RT-PCR | ||
| Weed plant species | |||||
| Amaranthus retroflexus | Amaranthaceae | 8/0 | − | 8/3 | + |
| Atriplex sagittata | Amaranthaceae | 6/0 | − | 7/0 | − |
| Chenopodium album | Amaranthaceae | 7/0 | − | 7/0 | − |
| Anthemis arvensis | Asteraceae | 8/8 | + | 8/8 | + |
| Chamomilla recutita | Asteraceae | 8/7 | + | 8/8 | + |
| Sonchus arvensis | Asteraceae | 8/0 | − | 8/0 | − |
| Galinsoga parviflora | Asteraceae | 8/0 | − | 8/0 | − |
| Capsella bursa-pastoris | Brassicaceae | 6/0 | NP | 6/0 | − |
| Stellaria media | Caryophyllaceae | 8/0 | − | 8/0 | − |
| Erodium cicutarium | Geraniaceae | 7/0 | − | 8/0 | − |
| Plantago major | Plantaginaceae | 7/0 | NP | 7/0 | − |
| Veronica agrestis | Scrophulariaceae | 7/0 | − | 7/0 | +c |
| Indicator plant species | |||||
| Gynura aurantiaca | Asteraceae | 4/4 | + | 4/4 | + |
| Lycopersicon esculentum | Solanaceae | 8/8 | + | 6/6 | + |
| Nicotiana benthamiana | Solanaceae | 8/8 | + | 8/8 | + |
See Materials and Methods for full scientific names.
For RT-PCR analyses, mixed RNA samples were analyzed for viroid at 25 days p.i.: +, positive, and −, negative. For the dot blot analyses, the number of plants inoculated/number of plants infected is shown for 25 days p.i. NP, not performed.
Only a weak signal was detected.
TABLE 2.
Detection of HSVd infection in inoculated weeds from hop fields
| Plant speciesa | Family | HSVd inoculum and detectionb
|
|||
|---|---|---|---|---|---|
| RNA
|
EcoRI cDNA fragments
|
||||
| Dot blot | RT-PCR | Dot blot | RT-PCR | ||
| Weed plant species | |||||
| Amarantus retroflexus | Amaranthaceae | 8/0 | − | 7/0 | +c |
| Atriplex nitens | Amaranthaceae | 8/0 | − | 8/0 | − |
| Artemisia vulgaris | Asteraceae | 8/0 | NP | 6/0 | − |
| Cirsium arvense | Asteraceae | 8/0 | − | 6/0 | − |
| Galinsoga ciliata | Asteraceae | 8/4 | + | 8/5 | + |
| Senecio vulgaris | Asteraceae | 8/0 | − | 8/0 | − |
| Sonchus arvensis | Asteraceae | 7/0 | − | 6/0 | − |
| Taraxacum officinale | Asteraceae | 8/0 | − | 6/0 | − |
| Capsella bursa-pastoris | Brassicaceae | 8/0 | − | 7/0 | − |
| Erysimum cheiranthoides | Brassicaceae | 8/0 | − | 8/0 | − |
| Epilobium parviflorum | Onagraceae | 8/0 | NP | 8/0 | − |
| Galium aparine | Rubiaceae | 8/0 | NP | 7/0 | − |
| Urtica urens | Urticaceae | 8/0 | − | 8/0 | − |
| Urtica dioica | Urticaceae | 8/0 | NP | 8/0 | − |
| Indicator plant species | |||||
| Gynura aurantiaca | Asteraceae | 4/4 | + | 4/4 | + |
| Humulus lupulus | Cannabaceae | 4/4 | + | 4/4 | + |
| Cucumis sativa | Cucurbitaceae | 8/8 | + | 8/8 | + |
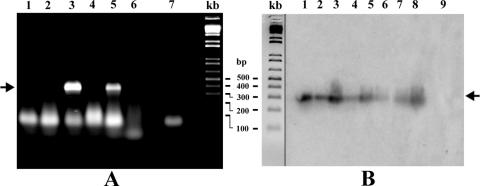
Biolistic inoculation with PSTVd RNA led to infection of the two related weed species chamomile (C. recutita) and corn chamomile (A. arvensis), both from the family Asteraceae. PSTVd was readily detectable by dot blot hybridization in these species as shown for corn chamomile in Fig. 1A. After inoculation with cDNA we detected a weak but specific dot blot hybridization signal in three of eight inoculated plants of redroot amaranth (A. retroflexus) from the family Amaranthaceae. The infection was confirmed by RT-PCR (Table 1 and Fig. 2A). A weak RT-PCR signal was also detected after cDNA inoculation of common speedwell (V. agrestis) from the family Scrophulariaceae (Table 1 and Fig. 2A). We did not detect any specific signals in other weed species tested for PSTVd even by RT-PCR (Table 1 and Fig. 2A), although 100% infections were detected for all indicator species.
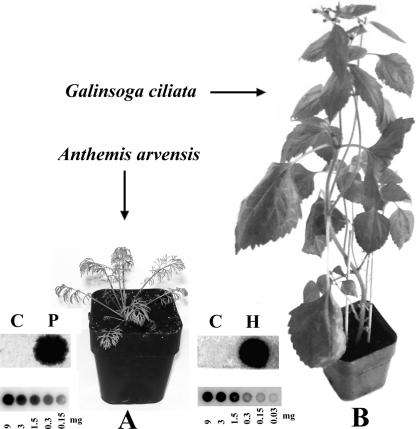
FIG. 1.
Examples of susceptible weeds infected with pospiviroids. (A) A. avensis, infected with PSTVd-RNA inoculum 25 days postinoculation; (B) G. ciliata infected with HSVd-RNA inoculum 32 days postinoculation. The dot blot signals corresponding to sample extracts from 9 mg of PSTVd (P)- or HSVd (H)-infected tissue (fresh mass) are shown for both examples. Mock-inoculated samples are designated by letter “C.” In smaller scale the dot blot hybridization signals are shown for a dilution series of a PSTVd sample from tomato (A) and of a HSVd sample from hop (B).
FIG. 2.
Examples of viroid detection in weed plants by RT-PCR. (A) RT-PCR analysis of PSTVd-infected weed species. Lanes: 1, G. parviflora; 2, S. media; 3, A. retroflexus; 4, A. sagittata; 5, V. agrestis; 6, E. cicutarium; 7, sample without RNA. No specific bands were detected in uninoculated plants. (B) RT-PCR, followed by molecular hybridization of HSVd samples (lanes 1 to 8) extracted from inoculated A. retroflexus 30 days postinfection. Position 9 represents a control sample from an uninoculated pant. In the center is shown a 1 KB Plus DNA Ladder (Life Technologies); the positions of PSTVd and HSVd-specific bands are indicated by the arrows.
It was of interest to verify, especially for chamomile, whether or not PSTVd can be transmitted by seeds. In our experiments, however, PSTVd-infected plants became sterile. Thus, this possibility has not been analyzed further.
The weeds we selected from hop gardens included most important field contaminants. We infected these weeds biolistically with an HSVdg RNA inoculum originating from Czech grapevines (see Materials and Methods). According to dot blot hybridization, of the selected weeds only hairy galinsoga (G. ciliata, family Asteraceae) was successfully infected (Table 2 and Fig. 1B). By biolistic infection using a cDNA inoculum, redroot amaranth (A. retroflexus) also became very weakly infected. The weak RT-PCR signal from redroot amaranth (Table 2) was verified in individual samples by molecular hybridization to a HSVd-specific probe (Fig. 2B). These results suggest a specific but very low level of HSVd in these species at 25 days postinfection. No specific signals were detected in other weed species, whereas 100% infections were detected in the indicator plant species hops, cucumber, and G. aurantiaca.
HSVd-infected G. ciliata could represent a dangerous “viroid transmitter” if it occurs in hop gardens. Note that this is especially true in relation to our previous research reporting a high HSVdg incidence in grapevines grown close to hop gardens in the Czech Republic (24). Thus, two additional factors were verified for this species: we checked, first of all, for infectivity of a natural RNA inoculum conventionally applied using Carborundum as an abrasive and, second, for possible seed transmissibility of HSVd. Twenty galinsoga plants were inoculated by mechanical inoculation, but none became infected. This suggests either a very low sensitivity or even insensitivity of this weed to conventional inoculation. This finding is consistent with the fact that even with the biolistic method the infectivity reached only ca. 50%. In addition, we found that HSVd is not transmissible through galinsoga seeds.
Analysis of viroid replication and levels in infected weed plants.
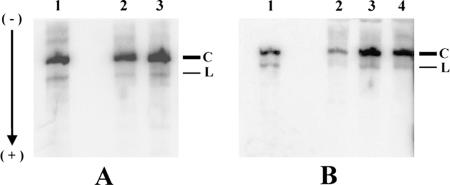
In our study we discovered several new experimental hosts of PSTVd and HSVd. Although in chamomile species and galinsoga the viroid levels were readily detectable by a dot blot procedure (for simplicity, we designated these weeds as “high viroid level species”), common speedwell and redroot amaranth viroids were unambiguously detected only by the much more sensitive RT-PCR method and after cDNA delivery (“low viroid level species”). In further experiments we analyzed RNA extracts from the “high viroid level” species in denaturing polyacrylamide gels to verify the presence of monomeric (+)-circular viroid RNA, which is indicative of a regular replication, processing, and circularization machinery in the host after inoculation with linear RNA or even cDNA. Indeed, monomeric (+)-circular viroid RNA was found in all “high viroid level” species as a band retarded in denaturing gels (Fig. 3). Ratios of radioactivity signals corresponding to monomeric circular to linear viroid RNAs were similar in PSTVd- and HSVd-infected indicator and weed species and ranged from 7.9 to 11.7. These results suggest normal PSTVd and HSVd replication in biolistically inoculated chamomile species and galinsoga, respectively. Viroid levels in redroot amaranth and common speedwell that are too low render the corresponding gel analysis impossible.
FIG. 3.
Presence of monomeric (+)-circular and linear viroid RNA in infected plants. (A) PSTVd from tomato (lane 1), C. reculita (lane 2), and A. arvensis (lane 3). Both weeds were inoculated with RNA. (B) HSVd from H. lupulus (lane 1), C. sativa (lane 2), and G. ciliata (lane 3) inoculated with DNA and G. ciliata inoculated with RNA (lane 4). Total RNA (15 μg from weeds or 10 μg from indicator plants) isolated using CONCERT plant RNA isolation reagent, treated with DNase, and purified by using a QIAGEN RNA purification kit were applied on 5.5% acrylamide gels containing 8 M urea and electrophoresed at 60°C. The nucleic acids were then electroblotted onto a nylon membrane and hybridized to viroid probes. C, monomeric circular positive-stranded viroid RNA; L, linear monomeric viroid RNA.
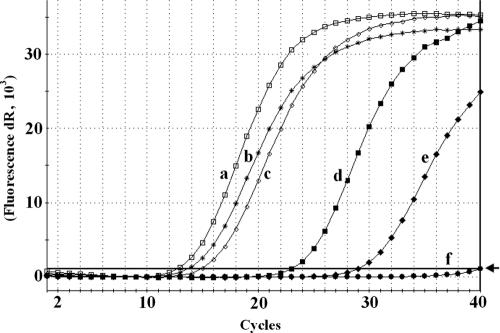
In order to estimate relative levels of PSTVd in various weed species in comparison to the indicator plant tomato, we developed a sensitive real-time PCR approach. The primers PCR-R and RT-R were chosen in positions where nucleotide changes are located neither in the original strains used for inoculation nor in the progeny strains (see Fig. 5). Amplification of the resulting specific 101-bp region was then quantified; a diluted sample of a clone with PSTVd sequence was used as standard for product quantification. As shown in Fig. 4, the levels of PSTVd in common speedwell and redroot amaranth were 105 and 103 lower than in tomato and reached 4.5 and 400 ag per 1 μg of total RNA, respectively, whereas in tomato 475 pg of PSTVd per 1 μg of RNA was detected. Levels of PSTVd in chamomile and corn chamomile were only 4.7 and 1.6 times lower than in tomato, respectively. These results suggest that the PSTVd propagation in chamomile species is quite comparable with that in tomato cv. Rutgers known as classical PSTVd host.
FIG. 5.
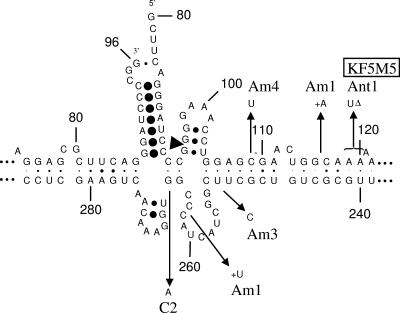
Localization of mutations in PSTVds from infected weeds. The secondary structure is based on consensus base-pairing probability of all PSTVd strains used for infection and resulting mutants found in the present study. The sequence of the PSTVd strain “intermediate” (9) is given. Nucleotide positions that differ in the strains and mutants from the “intermediate” sequence are marked by arrows. The names of PSTVd strains used for infecting weeds are boxed. Individual secondary structures of mutants and infecting, parental strains are available (see Fig. S1 at http://www.biophys.uni-duesseldorf.de/Matousek_JVI_2007/). Am1 to Am4, mutants isolated from Amaranthus retroflexus (redroot amaranth); Ant1 to Ant5, Anthemis arvensis (corn chamomile); C1 to C3, Chamomilla recutita (German chamomile); V1, Veronica arvensis (common speedwell). In gray are marked the domains in viroids (16). Primers used for cloning of mutants (PSTVdsI and PSTVdsII) and for quantification by real-time PCR (PCR-R and RT-R) are marked by dotted arrows.
FIG. 4.
Real-time PCR analysis of PSTVd levels in infected plant species. Total RNA was isolated from leaf tissues by using the CONCERT plant RNA purification reagent, followed by treatment with DNase and purification by using a QIAGEN RNeasy RNA cleaning protocol. RNA samples were subjected to RT and real-time PCR as described in Materials and Methods. Samples a to d correspond to templates from 0.2 μg of total RNA, samples e and f were from 1 μg of total RNA. Curves: a, Lycopersicon esculentum; b, Anthemis arvensis; c, Chamomilla recutita; d, Amaranthus retroflexus; e, Veronica agrestis; f, Erodium cicutarium. Amplified product in reactions a to e corresponded to the specific 101-bp fragment. No amplification product was detected in reaction f. The threshold level of fluorescence, as designated by the arrow, was set to a value of 1,028.
Analysis of viroid sequence variants propagating in infected weed species.
Sequence analyses of PSTVd and HSVd progenies propagated in infected weeds resulted in several new sequence variants. A spectrum of PSTVd progeny variants was found in the group of “high-level viroid” species, chamomile and corn chamomile, where PSTVd variants related to the various strains used for inoculation were detected (Fig. 5 and 6). None of the chamomile (C1 to C3) and corn chamomile (Ant1, -2, -4, and -5) sequences except for Ant3 was identical to one of the original strains; rather, they look like a patchwork from sequences of the mild KF5M5 and the lethal RG1 and KF440-2 strains. For example, Ant1 contains a C46A47 present in RG1 and KF440-2 and, present in KF5M5, a U instead of A120A121 and a CU instead of A310C311. However, all chamomile and corn chamomile sequences (except Ant3) contained additional, unique mutations (see below).
FIG. 6.
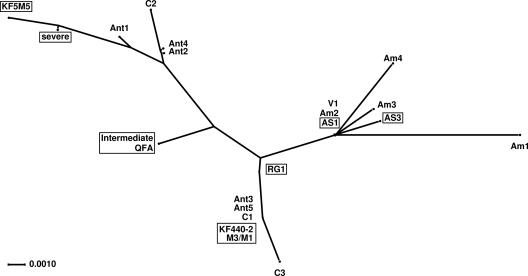
Tree of similarities between PSTVd strains used for infecting weeds and the resulting, mutated progeny strains. This “phylogenetic” tree was produced by SplitsTree4 (13) using a “sequence+structure” alignment (18). Distances were calculated by using the HKY85 model (11) with base frequencies of the alignment; the tree was computed by BioNJ (8). Despite the statistically low significance of the tree, no basically different tree or network was obtained by using other distance, tree, or network methods implemented in SplitsTree4.
In “low-level viroids” from A. retroflexus and V. argensis, biolistically inoculated with cDNA fragments, a more specific spectrum of sequence variants was detected. In V. argensis all clones were identical to the AS1 variant. In A. retroflexus all clones were related to the lethal AS1 sequence variant; that is, the sequence Am1 is identical to AS1, whereas the other PSTVd sequences from A. retroflexus deviate from AS1 only by additional mutations, which are unique according to a comparison with all PSTVd sequences known from the viroid database (30).
Some of the PSTVd mutants bore new specific mutations that were not characteristic for strains of the original inoculum. Examples are G→A base changes at positions 73 and 266 in PSTVd from chamomile and mutations U304A305→UAAU in PSTVd from corn chamomile (Fig. 5). In the PSTVd sequence population isolated from A. retroflexus new mutations were detected at the following positions: C109→U, U157→C, A219G220→UC, U251→C, C262CC264→CCUC, and G294→A. Some mutations are at positions that might influence the structure of the central conserved domain in processing configuration (1) (see Fig. 7); these are positions 109, 116/7, 251, and 263/4 in sequences of redroot amaranth and position 266 of chamomile. None of these mutations is, however, deleterious to the processing structure; two of them are even compensated for by base pair changes (G106:U251→G:C and C109:G249→U:G). These findings suggest some specificity in sequence changes connected to the process of viroid adaptation in infected weed species, as well as a strong sequence-selection process in the “low-level viroid” species A. retroflexus and V. agrestis.
FIG. 7.
CCR in processing configuration. A model of the conformation is shown that is critical for processing of PSTVd multimers to monomers (1). The site of 5′ cleavage is marked by an arrowhead. The size of dots connecting base pairs is proportional to consensus pairing probability in 11 pospiviroid sequences (35). Nucleotides differing in the mutants from the sequence of PSTVd “intermediate” are marked by arrows; for the names of the mutants, see Fig. 5.
Sequence analysis of viroids from the hop-field weed G. ciliata yielded a uniform result: all eight sequenced clones from this species are identical to a HSVdg variant described earlier (AC: E01844), which is strongly pathogenic for hops.
DISCUSSION
In the present study we analyzed experimental transmission of the two pospiviroids PSTVd and HSVd to specific weed plants selected from potato fields and hop gardens. For this purpose we used an efficient biolistic method (25, 26) to transfer viroid populations. As a result of this analysis, several plant species growing in corresponding agrobiotopes as weeds (chamomile, corn chamomile, redroot amaranth, common speedwell, and galinsoga) were found to be experimental hosts having the potential to propagate viroids. Consequently, these plants represent a potential risk for cultured plants if infected accidentally. One has to consider, however, that this risk depends on several factors, such as sensitivity to infection, viroid levels, seed transmissibility and last, but not least, viroid changes associated with possibly fast evolution and adaptation. “High-viroid-level” species such as chamomile and corn chamomile propagating PSTVd and galinsoga propagating HSVd are widely spread field contaminants. Viroid levels and replication in these plants are similar to that in known indicators. In this respect especially cultured chamomile is a crop with a potential risk to become infected. On the other hand, the thread-like, pinnate morphology, and anatomy of leaves of chamomile and corn chamomile could form some barrier for a Carborundum-based inoculation at least at the stage of seedlings and young plants; that is, the mechanically injured leaves tend to dry, thus preventing an infection. In addition, we found infected wild chamomile species to be sterile. This sterility is probably associated with viroid pathogenesis in this species. A more detailed analysis of PSTVd-induced pathogenesis in cultured chamomile is in progress. The important factor that decreases the risk of HSVd infections in galinsoga is, according to our results, lower sensitivity to inoculation and the fact that HSVd was not seed transmissible in this weed species. It is important to note that all of these weed species are from the family Asteraceae like one of the indicator plants, G. aurantiaca, that is a known host for both pospiviroids (2, 10). No visible pathogenesis symptoms associated with PSTVd-infected V. agrestis and A. retroflexus and HSVd-infected A. retroflexus and G. ciliata were observed.
“Low-viroid-level” species such as redroot amaranth and common speedwell represent a phenomenon that is in accordance with our previous studies describing PSTVd propagation in some Brassica species (25) or HLVd propagation in Solanaceous species after experimental transmission (21). Low levels of HLVd were also detected by RT-PCR in naturally infected Urtica dioica trichomes (17) and in leaves in our experiments (unpublished). In such species viroid populations were maintained at levels hardly detectable by molecular hybridization methods. In addition, low viroid levels were detectable in weed species analyzed in the present study only after inoculation with infectious viroid cDNAs, whereas no signal was detected after inoculation with RNA. This suggests high insensitivity to infection with natural viroids. It is interesting that redroot amaranth and common speedwell are from families the Amaranthaceae and Scolpulariaceae, respectively, and that other PSTVd hosts from these families are in the list of PSTVd-susceptible plants (33; for a review, see reference 2).
The mechanism maintaining viroid propagation at low levels is not known. No natural resistance against viroid has been reported (for a review, see reference 10). For instance, Singh (34), who tested S. berthaultii as a possible source of resistance, revealed clones resistant to mechanical inoculation. However, these clones become infected with low levels of viroid after PSTVd transmission through grafting (34). It is believed that plant resistance to molecular pathogens is at least partly mediated by the branched pathways of posttranscriptional gene silencing (for reviews, see references 37 and 38). Although the characteristic RNA cleavage products of about 22 to 25 nt appearing during the viroid propagation cycle (14, 19, 20, 23, 29) are indicative of viroid-mediated induction of gene silencing, the rod-like viroid RNA structure is not an effective substrate for degradation by RNA-induced silencing complex (15); this structural feature probably developed as viroid adaptation to escape the silencing machinery (15, 39). Whether or not a silencing mechanism or rather an inefficient replication and/or transportation mechanisms lead to low-level viroid populations in some species remains to be clarified.
New sequence variants of PSTVd were found in experimentally infected weed species. The positions of unique new mutations were predominantly located in but not restricted to the pathogenicity domain, as in the case of “thermomutants” described in our previous work (26, 27). Some mutations detected in PSTVd from redroot amaranth and chamomile were localized in the central conserved region similarly to those detected in the low-level population of PSTVd from A. thaliana (26). According to structural calculations, these mutations have no major influence on the thermodynamic stability of PSTVd's native structure (Fig. 5 and Fig. S1 at http://www.biophys.uni-duesseldorf.de/Matousek_JVI_2007/) or processing conformation (Fig. 7). Our results rather demonstrate that PSTVd replication in new experimental hosts is associated with viroid mutagenesis that may be important for subsequent viroid adaptation to new host species. Simultaneously, this adaptation was demonstrated with biolistically transferred PSTVd and HSVd populations, where differential selection occurred in various hosts, eliminating or overgrowing some of the sequences originally included in the inoculum. The sequence variations, however, would not prohibit an easy (re)infection of economic plants.
Acknowledgments
We thank Milena Matoušková, Helena Matoušková, and Olga Horáková from the Biology Centre of the ASCR, v.v.i. Institute of Plant Molecular Biology, České Budějovice, Czech Republic, for their help and excellent technical assistance and Bernd Esters (Institute of Physical Biology, Heinrich-Heine-Universität, Düsseldorf, Germany) for fruitful discussions and excellent technical support.
The study was supported by grants AS CR 1QS500510558 and AV0Z50510513, as well as by Deutsche Forschungsgemeinschaft grant RI252/19.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Baumstark, T., A. R. Schröder, and D. Riesner. 1997. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diener, T. O. (ed.). 1979. Viroids and viroid diseases. Wiley, New York, NY.
- 3.Diener, T. O. (ed.). 1987. The viroids. Plenum Press, Inc., New York, NY.
- 4.Diener, T. O. 1995. Origin and evolution of viroids and viroid-like satellite RNAs. Virus Genes 11:119-131. [DOI] [PubMed] [Google Scholar]
- 5.Eigen, M. 1993. Viral quasispecies. Sci. Am. 269:42-49. [DOI] [PubMed] [Google Scholar]
- 6.Flores, R., C. Hernández, A. E. Martínez de Alba, J. A. Daròs, and F. Di Serio. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 7.Flores, R., J. W. Randles, M. Bar-Joseph, and T. O. Diener. 1998. A proposed scheme for viroid classification and nomenclature. Arch. Virol. 143:623-629. [DOI] [PubMed] [Google Scholar]
- 8.Gascuel, O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 9.Gross, H. J., H. Domdey, C. Lossow, P. Jank, M. Raba, H. Alberty, and H. L. Sänger. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273:203-208. [DOI] [PubMed] [Google Scholar]
- 10.Hadidi, A., R. Flores, J. W. Randles, and J. S. Semancik (ed.). 2003. Viroids. CSIRO Publishing, Collingwood, Victoria, Australia.
- 11.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 12.Hofacker, I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31:3429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 14.Itaya, A., A. Folimonov, Y. Matsuda, R. S. Nelson, and B. Ding. 2001. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant-Microbe Interact. 14:1332-1334. [DOI] [PubMed] [Google Scholar]
- 15.Itaya, A., X. Zhong, R. Bundschuh, Y. Qi, Y. Wang, R. Takeda, A. R. Harris, C. Molina, R. S. Nelson, and B. Ding. 2007. A structured viroid RNA is substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81:2980-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keese, P., and R. H. Symons. 1985. Domains in viroids: evidence of intermolecular RNA rearrangement and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 82:4582-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knabel, S., H. L. Seigner, and P. R. Wallnöfer. 1999. Detection of hop latent viroid (HLVd) using the polymerase chain reaction (PCR). Gesunde Pflanzen. 51:243-249. [Google Scholar]
- 18.Lück, R., S. Gräf, and G. Steger. 1999. ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res. 27:4208-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markarian, N., H. W. Li, S. W. Ding, and J. S. Semancik. 2004. RNA silencing as related to viroid induced symptom expression. Arch. Virol. 149:397-406. [DOI] [PubMed] [Google Scholar]
- 20.Martínez de Alba, A. E., R. Flores, and C. Hernández. 2002. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J. Virol. 76:13094-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matoušek, J. 2003. Hop latent viroid (HLVd) microevolution: an experimental transmission of HLVd “thermomutants” to solanaceous species. Biologia Plantarum 46:607-610. [Google Scholar]
- 22.Matoušek, J., V. Junker, L. Vrba, J. Schubert, J. Patzak, and G. Steger. 1999. Molecular characterization and genome organization of 7SL RNA genes from hop (Humulus lupulus L.). Gene 239:173-183. [DOI] [PubMed] [Google Scholar]
- 23.Matoušek, J., P. Kozlová, L. Orctová, A. Schmitz, K. Pešina, O. Bannach, N. Diermann, G. Steger, and D. Riesner. 2007. Accumulation of viroid-specific small RNAs and increase of nucleolytic activities linked to viroid-caused pathogenesis. Biol. Chem. 388:1-13. [DOI] [PubMed] [Google Scholar]
- 24.Matoušek, J., L. Orctová, J. Patzak, P. Svoboda, and I. Ludvíková. 2003. Molecular sampling of hop stunt viroid (HSVd) from grapevines in hop production area in the Czech Republic and hop protection. Plant Soil Environ. 49:168-175. [Google Scholar]
- 25.Matoušek, J., L. Orctová, G. Steger, and D. Riesner. 2004. Biolistic inoculation of plants with viroid nucleic acids. J. Virol. Methods 122:153-164. [DOI] [PubMed] [Google Scholar]
- 26.Matoušek, J., L. Orctová, G. Steger, J. Škopek, M. Moors, P. Dědič, and D. Riesner. 2004. Analysis of thermal stress-mediated PSTVd variation and biolistic inoculation of progeny of viroid “thermomutants” to tomato and Brassica species. Virology 323:9-23. [DOI] [PubMed] [Google Scholar]
- 27.Matoušek, J., J. Patzak, L. Orctová, J. Schubert, L. Vrba, G. Steger, and D. Riesner. 2001. The variability of hop latent viroid as induced upon heat treatment. Virology 287:349-358. [DOI] [PubMed] [Google Scholar]
- 28.Matoušek, J., A. R. W. Schröder, L. Trněná, M. Reimers, T. Baumstark, P. Dědič, J. Vlasák, I. Becker, F. Kreuzaler, M. Fladung, and D. Riesner. 1994. Inhibition of viroid infection by antisense RNA expression in transgenic plants. Biol. Chem. Hoppe-Seyler 375:765-777. [DOI] [PubMed] [Google Scholar]
- 29.Papaefthimiou, I., J. A. Hamilton, M. A. Denti, D. C. Baulcombe, M. Tsagris, and M. Tabler. 2001. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of posttranscriptional gene silencing. Nucleic Acids Res. 29:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocheleau, L., and M. Pelchat. 2006. The Subviral RNA Database: a toolbox for viroids, the hepatitis delta virus and satellite RNAs research. BMC Microbiol. 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sano, T., R. Mimura, and K. Ohshima. 2001. Phylogenetic analysis of hop and grapevine isolates of hop stunt viroid supports a grapevine origin for hop stunt disease. Virus Genes 22:53-59. [DOI] [PubMed] [Google Scholar]
- 32.Schnölzer, M., B. Haas, K. Ramm, H. Hofmann, and H. L. Sänger. 1985. Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 4:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh, R. P. 1973. Experimental host range of the potato spindle tuber “virus.” Am. Potato J. 50:111-123. [Google Scholar]
- 34.Singh, R. P. 1985. Clones of Solanum berthaultii resistant to potato spindle tuber viroid. Phytopathology 75:1432-1434. [Google Scholar]
- 35.Steger, G., and D. Riesner. 2003. Properties of viroids: molecular characteristics, p. 15-29. In A. Hadidi, R. Flores, J. W. Randles, and J. S. Semancik (ed.), Viroids. CSIRO Publishing, Collingwood, Victoria, Australia.
- 36.Tabler, M., and M. Tsagris. 2004. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 9:339-348. [DOI] [PubMed] [Google Scholar]
- 37.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 38.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 39.Wang, M. B., X. Y. Bian, L. M. Wu, L. X. Liu, N. A. Smith, D. Isenegger, R. M. Wu, C. Masuta, V. B. Vance, J. M. Watson, A. Rezaian, E. S. Dennis, and P. M. Waterhouse. 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 101:3275-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]