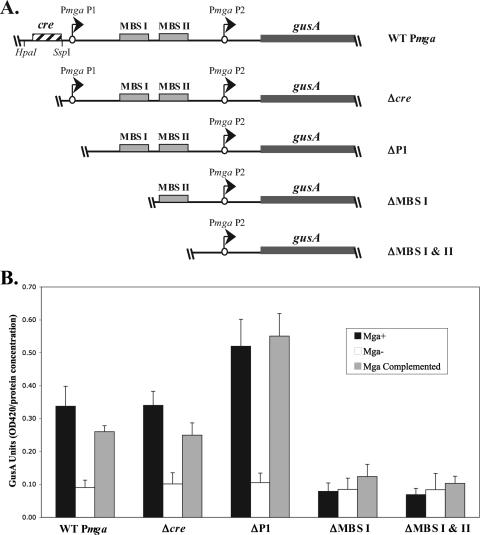
FIG. 3.
VIT GusA transcriptional reporter assay for deletion analysis of Pmga. (A) Schematic representation of gusA transcriptional reporters in the chromosomal VIT locus of the M6 GAS corresponding to wild-type Pmga (KSM310), Δcre (KSM435), ΔP1 (KSM427), ΔMBS I (KSM428), and ΔMBS I & II (KSM429). The starts of transcription for mga (circles with arrows), the gusA gene (thick line), MBSs (solid boxes), putative cre (striped box), and relevant restriction sites are shown. (B) GusA assays for the Pmga-gusA constructs depicted in panel A inserted into the VIT locus of the wild-type JRS4-derived M6 GAS strain RTG229 (black bars), isogenic mga-inactivated strain KSM150Lg (open bars), and mga-inactivated KSM150Lg complemented with the Pspac-mga plasmid pKSM162 (shaded bars). Data are reported in GusA units (OD420/concentration of total protein [in micrograms per microliter]) and represent an average of the results from at least three independent experiments. The error bars express the standard deviations for each strain measured.