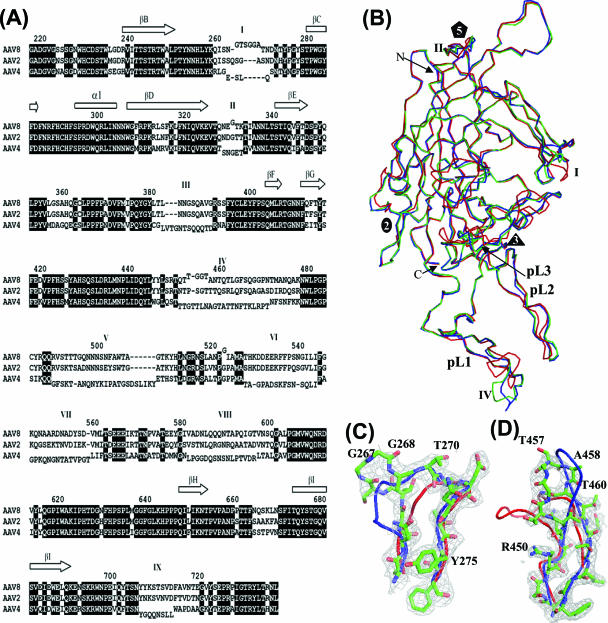
FIG. 2.
Superimposition of AAV structures. (A) Sequence/structure alignment of the AAV2, AAV4, and AAV8 VP3 amino acids. The core β-strands forming the β-barrel are represented as arrows at the top of the alignment, and variable regions I to IX (as defined in reference 18) are indicated above the sequence. VP regions that differ structurally between the viruses are offset above (for AAV8) and below (for AAV4) the alignment. The residue numbering above the alignment is based on AAV8 VP1 and refers to the center residue. (B) Superimposition of C-α atoms of AAV2 (blue), AAV4 (red), and AAV8 (green). The three loops at which AAV8 differs from AAV2 are located at variable regions I (residues 263 to 271), II (residues 329 to 333), and IV (residues 452 to 471). (C) Superimposition of the variable region I surface loops of AAV2 (blue), AAV4 (red), and AAV8 (green) and the 2Fo-Fc map of AAV8 (gray mesh). (D) Superimposition of the variable region IV loops and the 2Fo-Fc map of AAV8. The color scheme follows that of panel C.