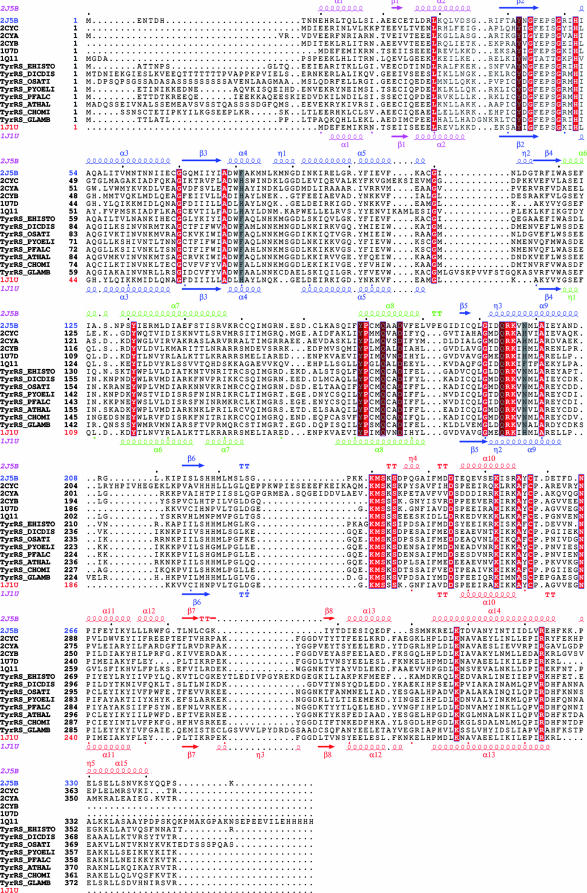
FIG. 3.
Structure-based alignment of TyrRSs. TyrRSapm (2J5B) was aligned with eukaryal (1Q11, human, core structure) and archaeal (2CYC, P. horikoshii; 2CYA, Aeropyrum pernix; 2CYB, Archaeoglobus fulgidus; 1J1U and 1U7D, M. jannaschii complex and apo form) structures. The closest TyrRSapm homologues from protozoa and plants are also included (EHISTO, Entamoeba histolytica; DICDIS, Dictyostelium discoideum; OSATI, Oryza sativa; PYOELI, Plasmodium yoelii; PFALC, Plasmodium falciparum; ATHAL, Arabidopsis thaliana; CHOMI, Cryptosporidium hominis; GLAMB, Giardia lamblia). The secondary-structure elements of TyrRSapm and M. jannaschii are, respectively, indicated above and below the multiple alignment. The N-terminal, Rossmann fold, CP1, and C-terminal domains are colored in pink, blue, green, and red, respectively. Strictly conserved residues are boxed in red. Residues involved in tyrosine binding (Fig. 2) are highlighted in gray. This alignment was produced with 3DCoffee (http://www.igs.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi) (48), and the figure was produced with ESPript (31).