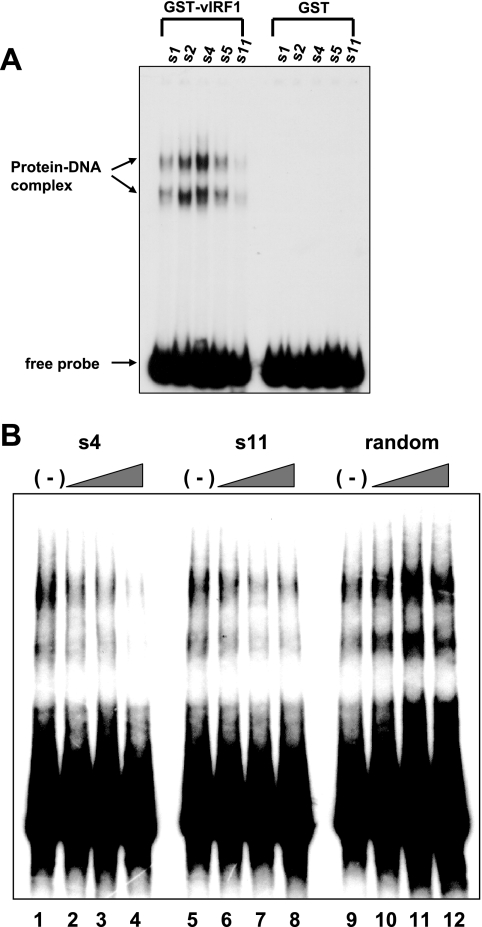
FIG. 2.
Characterization of vIRF1-binding sites. (A) Gel shift assays with the selected vIRF1-binding sites. Gel shift assays were performed with selected DNA (s1, s2, s4, s5, and s11) sequences and GST-vIRF1 (lanes 1 to 5) or GST protein (lanes 6 to 10). Protein-DNA complexes are specified using arrows. (B) Competition assays were performed by preincubation with selected unlabeled vIRF1-binding sites. Labeled s5 probe was used in each gel shift assay. Unlabeled s4 (lanes 1 to 4), s11 (lanes 5 to 8), and random oligonucleotides (lanes 9 to 12) were used. Twofold (lanes 2, 6, and 11), fivefold (lanes 3, 7, and 12), and 10-fold (lanes 4, 8, and 12) molar excesses of unlabeled probes were used, respectively.