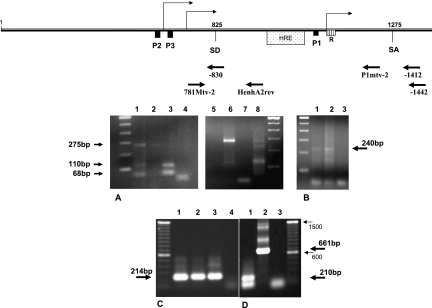
FIG. 4.
Determination of the start of transcription for the novel MMTV P3 promoter. (A) 5′ RACE. DNase I-treated total RNA extract from N2AnE5 or NMuMG cells was reverse transcribed with a primer specific for env, and 5′ poly(A)-tailed, purified cDNA was used as a template for the first PCR amplification together with the gag-specific reverse primer −1412 and the 5′ RACE oligo(dT) anchor primer. Nested PCR was then performed using the anchor primer and primer −830 for the detection of the transcription start from the P3 promoter or using primer P1mtv-2 for the detection of the transcription start from the P1 promoter. PCR products were gel purified and subcloned for sequencing analysis. Lanes 1 to 4 show cells that were not treated with dexamethasone prior to RNA extraction. Lane 1, 5′ RACE PCR products using P1mtv2 from N2AnE5 cDNA; lane 2, 5′ RACE PCR products using P1mtv2 from NMuMG cDNA; lane 3, 5′ RACE PCR products using −830 from N2AnE5 cDNA; lane 4, 5′ RACE PCR products using −830 from NMuMG cDNA. Lanes 5 to 8 show cells that were treated with dexamethasone prior to RNA extraction. Lane 5, 5′ RACE PCR products using P1mtv2 from NMuMG cDNA; lane 6, 5′ RACE PCR products using P1mtv2 from N2AnE5 cDNA; lane 7, 5′ RACE PCR products using −830 from NMuMG cDNA; lane 8, 5′ RACE PCR products using −830 from N2AnE5 cDNA. (B) 5′ RLM-RACE. mRNA was dephosphorylated, the cap structure was removed, RNA was then ligated into the GeneRacer RNA oligonucleotide and reverse transcribed with the gag-specific primer −1442, and nested PCR was performed with primer −1412 and the 5′ GeneRacer primer. Lane 1, GeneRacer PCR products from GR mRNA; lane 2, GeneRacer PCR products from N2AnE5; lane 3, PCR negative control. GeneRacer PCR products were then gel purified before subcloning and sequencing. (C and D) Splicing. Total RNAs from MMTV producer GR cells were first reverse transcribed using specific reverse primer −1442, and RNase H treatment was performed, followed by PCR analysis. (C) Nonspliced transcripts. Primers 781Mtv-2 and HenhA2rev were used. The templates were reverse-transcribed RNAs from GR cells treated with dexamethasone (GR+dex RT RNAs) (lane 1), GR−dex RT RNAs (lane 2), and pGR102 DNA (lane 3). Lane 4, no-template control. (D) Spliced transcripts. PCR was performed with primers 781Mtv-2 and −1412. The templates were GR−dex RT RNAs (lane 1) and pGR102 DNA indicating the genomic size of a nonspliced fragment (lane 2). Lane 3 is a no-template PCR control. The 214-bp- and 210-bp-long DNA products were purified from gels shown in C and D. The sequence of the 210-bp fragment reveals a splicing event that occurred between SD825 and SA1275.