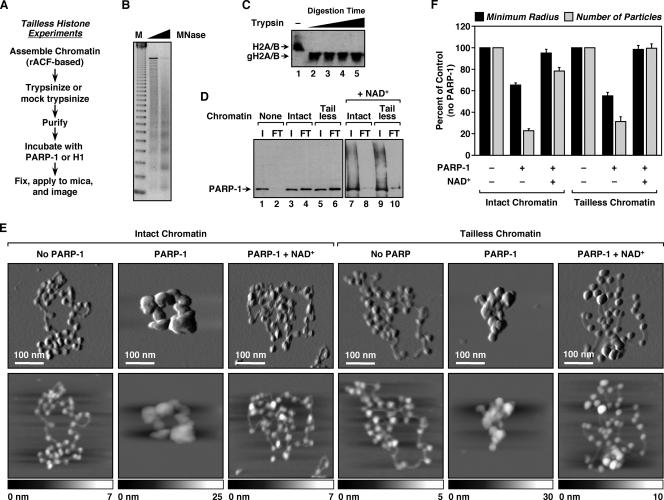
FIG. 3.
Histone tails are dispensable for PARP-1-mediated chromatin compaction. (A) Overview of the preparation of ACF-assembled tailless chromatin samples for AFM imaging (see Materials and Methods). (B) Agarose gel electrophoresis of MNase-digested chromatin assembled using recombinant ACF and pERE (see Materials and Methods). M, 123-base pair ladder. (C) Western blot for H2A and H2B illustrating the effective removal of histone tails by trypsin treatment. The digestion condition shown in lane 2 (i.e., complete, not excessive, digestion) was used for all studies with tailless chromatin. gH2A/B, globular (i.e., tailless) H2A and H2B. (D) PARP-1 chromatin binding assays. (Lanes 1 to 6) PARP-1 was subjected to small-scale size exclusion chromatography in the absence or presence of intact or tailless chromatin to assess its chromatin binding ability (see Materials and Methods). Two percent of the input (I) and 10% of the FT were analyzed by Western blotting for PARP-1. In this assay, unbound PARP-1 has a high residence time on the resin and thus is not evident in the FT. In contrast, chromatin-bound PARP-1 passes through the column with the FT. (Lanes 7 to 10) The effect of NAD+ on PARP-1 binding to chromatin was assayed in a similar manner. (E) AFM images of intact and tailless pFASTbac1-cDNA chromatin molecules alone, with a saturating amount of PARP-1, or with a saturating amount of PARP-1 plus NAD+, as indicated. Scan probe oscillation amplitude images (top) and topographical images (bottom) are as described for Fig. 1C. (F) Quantification of AFM images like those shown in panel E, as described in the legend to Fig. 1D. All determinations are based on at least 25 molecules derived from two independent preparations of chromatin. Each bar is the mean plus standard error of the mean.