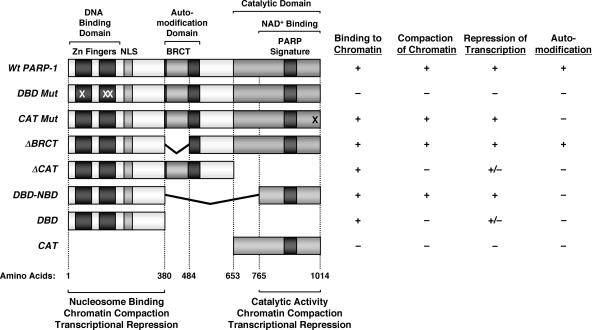
FIG. 8.
Summary of binding, chromatin compaction, transcriptional repression, and automodification characteristics of PARP-1 mutants. (Left) Schematic diagrams showing the structural and functional domains of PARP-1, as well as a set of PARP-1 deletion and point mutants. The locations of the point mutations in DBD mut (C21G, C125G, and L139P) and CAT mut (E988K) are marked with Xs. The amino acid start points and end points for the mutants are shown. NLS, nuclear localization signal. (Right) The PARP-1 deletion mutants shown were expressed in E. coli and purified by nickel-NTA affinity chromatography (see Materials and Methods). The purified proteins were screened for four different activities, as indicated: (i) binding to chromatin, as determined by in vitro chromatin binding assays (Fig. 4B and 6B); (ii) compaction of chromatin, as determined by AFM imaging (Fig. 4C and D and 6C and D); (iii) repression of transcription, as determined by in vitro transcription assays (Fig. 7D); and (iv) automodification, as determined by an in vitro PARylation assay in the presence of [32P]NAD+ (Fig. 5B). The data for DBD mut and CAT mut are from Kim et al. (15). Activity key: + indicates at least 75% of wild-type PARP-1 activity, − indicates less than 20% of wild-type PARP-1 activity, and +/− indicates ∼40 to 60% of wild-type PARP-1 activity.