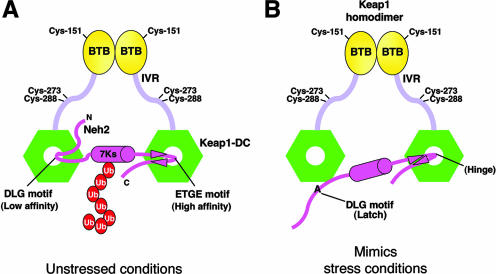
FIG. 5.
Alanine mutation in the DLG motif mimics oxidative stress conditions. (A) Schematic diagram showing the two-site substrate recognition model. Keap1 is composed of BTB (yellow ovals), intervening region (IVR) (light violet bars), and DGR/CTR (Keap1-DC; green hexagons) domains. Some of the important reactive cysteines are labeled. Keap1 homodimer binds with one Neh2 domain (magenta hockey stick) of Nrf2. The middle magenta cylinder represents the α-helix, and the magenta arrowheads represent the β-strands of Neh2. All seven lysines (7Ks) of Neh2 are located within the α-helical region. The multiple ubiquitin (Ub) units are shown as red ovals. Neh2 binds to Keap1-DC via both the ETGE and DLG motifs under unstressed conditions. This complex configuration facilitates efficient ubiquitination of the lysines in the α-helix of Neh2. (B) Alanine mutations in the DLG motif reduce its affinity to Keap1 as well as debilitate Keap1-mediated ubiquitination of Nrf2, which mimics conditions of oxidative stress. The DLG motif, therefore, acts as a latch to lock and unlock the lysines of Neh2 in an appropriate spatial orientation for targeting ubiquitin transfer by the E2 enzyme in different cellular redox states.