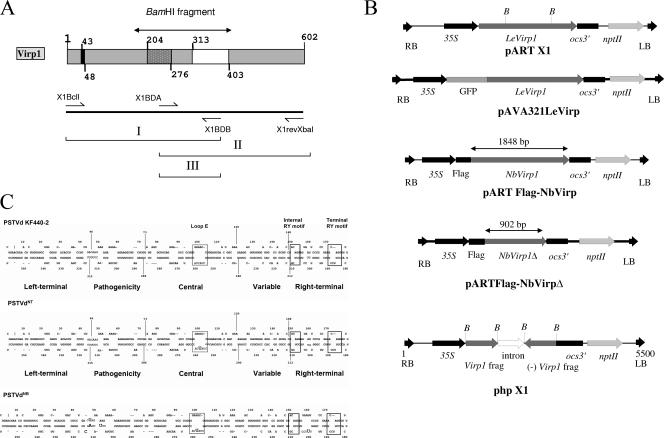
FIG. 1.
(A and B) Schematic representation of the tomato Virp1 protein (A, top); the location of the primers used for the RT-PCR (A, bottom); and the constructs used (B) for overexpression of Virp1, GFP-Virp1 fusion, FLAG-NbVirp1, FLAG-NbVirp1Δ, and suppression of Virp1 (from top to bottom, respectively) in N. benthamiana and N. tabacum transgenic plants. (A) (Top) Virp1 contains a nuclear localization signal (amino acids 43 to 48), a bromodomain (amino acids 204 to 276), and a potential RNA binding domain (amino acids 313 to 403). (B) nptII, selection gene; ocs3′, ocs terminator sequence; LB and RB, T-fragment left and right border sequences, respectively; 35S, cauliflower mosaic virus 35S promoter; B, BamHI sites delimiting the bromodomain. The truncated FLAG-NbVirp1Δ contains the first 902 bp of the NbVirp1 sequence. (C) Comparison of PSTVd strains KF 440-2, NT, and NB, showing the relative locations of the sequence differences between these three strains with respect to the loop E (dotted-line boxes) and RY motifs (solid-line boxes). In addition to a C259U substitution in the loop E motif of PSTVd-NT (shown by an oversized letter), strain PSTVd-NB also contains five additional mutations (shown by oversized letters).