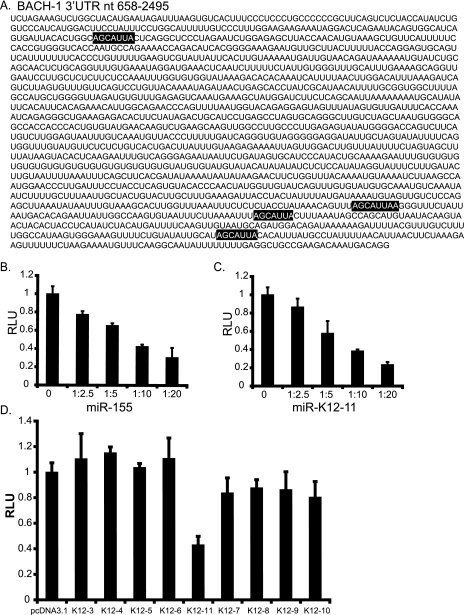
FIG. 3.
BACH-1 is targeted by miR-K12-11 and miR-155. (A) The BACH-1 3′UTR contains four seed match sites (highlighted) based on three target prediction programs (miRanda, PicTar, and TargetScan). (B and C) A region of the BACH-1 3′UTR (nt 658 to 2495) encompassing the predicted miRNA binding sites was cloned downstream of luciferase (pGL3-BACH1). 293 cells were cotransfected with 40 ng pGL3-BACH-1 and increasing amounts (100, 200, 400, and 800 ng) of pmiR-155 or pmiR-K12-11. Ratios on the x axis indicate the amount of miRNA vector to reporter. pcDNA3.1 was used as filler. Relative light units (RLU) are normalized to total protein determined by BCA (Pierce). (D) Additional KSHV miRNAs do not target the BACH-1 3′UTR. 293 cells were cotransfected with 40 ng pGL3-BACH-1, 10 ng pEF-RL (Renilla), and 800 ng of either pcDNA3.1 or KSHV miRNA expression vector. Light units are normalized to Renilla luciferase. For B to D, average data from three independent transfections are shown.