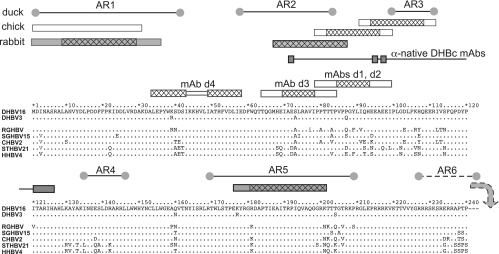
FIG. 3.
Epitope locations in the primary sequence of DHBc aligned with other avihepadnaviral core proteins. For DHBV16 core protein, the full sequence is shown, except for the C-terminal residues 241 to 262. Identical amino acids in the other proteins are designated by dots. The boxes above the sequence depict, from bottom to top, the epitopes of the anti-denatured DHBc MAbs and of polyclonal rabbit (dark) and chicken (light) anti-DHBc sera. For PepScan-mapped MAb epitopes, the cross-hatched areas refer to the common core sequence of the recognized peptides. For the polyclonal sera, cross-hatching indicates peptides that gave stronger signals. At the top, the previously reported linear antigenic regions (AR1 to AR6) recognized by duck antisera (18) are shown. AR6 is downstream of DBHc aa 240. The connected rectangles labeled “anti-native DHBc MAbs” show the sites where sequence modifications reduced recognition by these MAbs but not particle formation.