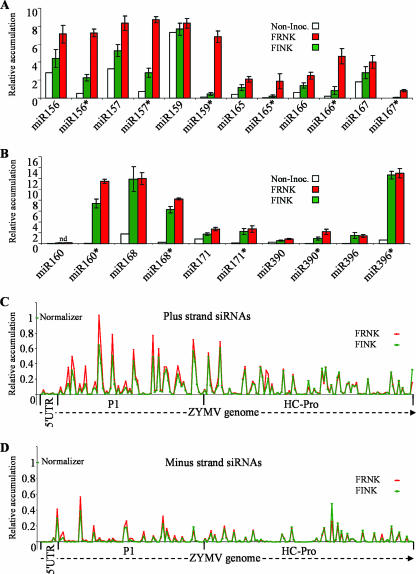
FIG. 2.
smRNAs of ZYMV-infected squash were quantified by microarray analysis at 5 dpi. The average expression levels of selected miRNAs (miR) and miRNA*s (miR*) that were quantified by using a comprehensive smRNA microarray assay (see Fig. S1 in the supplemental material for sequences shown in Fig. 2A and B). Noninoculated squash plants (Non-Inoc.) were compared to attenuated ZYMVFINK (FINK)- and severe ZYMVFRNK (FRNK)-infected plants. This graphic is arbitrarily divided to present two charts of miRNA/miRNA* pairs. For the pairs presented in panel A, the concentration of FRNK miRNA* is more than threefold greater than the concentration of FINK miRNA*. For the pairs presented in panel B, the concentration of FRNK miRNA* is less than threefold greater than the concentration of FINK miRNA*. The signals were normalized in-chip to a spike-in control RNA, and chip signals were subsequently normalized to each other. The smRNAs that were labeled and hybridized were from pools of seven to nine plants for each treatment. Error bars indicate the SEMs of three independent biological repeats. The relative accumulations of smRNAs derived from the plus (C) and minus (D) strands of the ZYMV genome in ZYMVFRNK (FRNK)- and ZYMVFINK (FINK)-infected leaves were measured. Viral siRNAs were assayed using tiled 25-nt probes on a representative microarray, as described above, that were tiled with a 13-bp overlap. Data shown are for 193 consecutive plus-strand probes and their complementary minus-strand probes that represent the first 2,437 nt, which encode the 5′ UTR protein, P1, and HC-Pro. For clarity in panels C and D, only one of three biological replicates is shown.