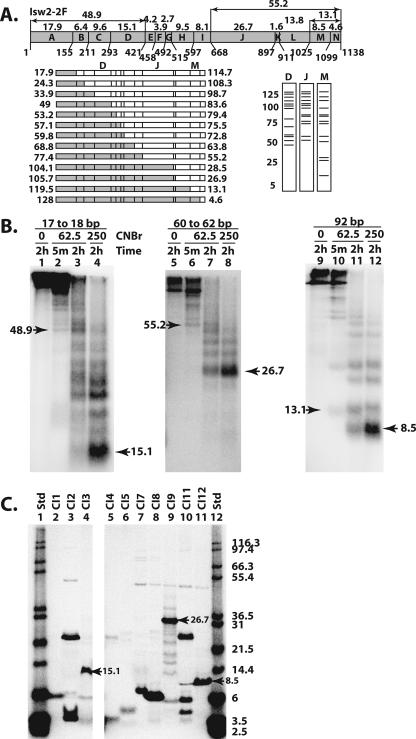
FIG. 3.
The DEXD, HAND, and SLIDE domains were cross-linked to nucleosomal and extranucleosomal DNA. (A) The methionine sites (numbers below the schematic) and predicted molecular masses (numbers above the schematic) of the proteolytic fragments are shown for Isw2. The set of fragments obtained with single-hit CNBr digestion are shown below the Isw2 schematic on the left, with two fragments created from each single hit depicted as either a shaded or an open box. Below on the right are shown the predicted radiolabeled fragments as resolved by SDS-PAGE and obtained from cross-linking in three different regions of Isw2, labeled D, J, and M. (B) Photoaffinity-labeled Isw2 was digested with CNBr at the indicated concentrations (mM) and incubation times, followed by analysis by 10% BIS-Tris SDS-PAGE. The apparent molecular masses, in kDa, of the released bands were calculated as described in Materials and Methods. The molecular masses for the smallest bands released by limited and extensive CNBr digestion are indicated by arrowheads. (C) Peptides corresponding to fragments of Isw2 that could be expected from complete CNBr digestion (Table 1) were synthesized and were analyzed by 10% BIS-Tris SDS-PAGE. Arrowheads highlight the markers that show migrations similar to those of the bands observed in CNBr peptide-mapping experiments. The molecular masses in kDa of 125I-labeled Mark12 protein standards (Std) are indicated.