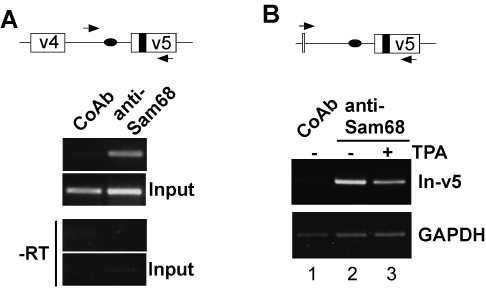
Figure 4. Signaling-induced interference with Sam68 binding to pre-mRNA in vivo.
(A) Detection of Sam68 binding to an endogenous CD44 pre-mRNA fragment (spanning the v5 exon and 100 bp upstream intron sequence) in LB17 lymphoma cells by RNP immunoprecipitations. A control antibody (CoAb) or an anti-Sam68 antibody were used for precipitations. Lower panels show amplifications from the lysate used as input for the precipitations. Bands correspond to the expected size of 228 bp. Positions of primers used for amplification are indicated as arrowheads in scheme on top; Sam68 binding sites (black oval and box, respectively), CD44 exons v4 and v5 (open boxes), intron (line). –RT represents reactions without reverse transcriptase. (B) RNP immunoprecipitations with either a control antibody (CoAb) or an anti-Sam68 antibody from lysates of LB17 lymphoma cells that were transfected with the pETv5 minigene construct [23]. The cells were either left untreated (−) or treated with 40 ng/ml of 12-o-tetradecanoylphorbol-13-acetate (TPA) for 10 min. A fragment from the minigene pre-mRNA spanning the v5 exon and 450 bp upstream intron sequence (In-v5) was amplified using the primers indicated (scheme on top; symbols as in panel A). Specific amplification of the minigene pre-mRNA is achieved by the forward primer that is complementary to the deletion junction of the v4 exon in the construct. To check for potential differential loss of material during the precipitation and washing procedure, we monitored the amounts of GAPDH mRNA unspecifically bound to the antibody and/or the agarose beads (lower panel). Bands correspond to the expected sizes of 600 bp (In-v5) and 278 bp (GAPDH). All PCR amplifications were in linear phase as verified with different amounts of cDNA.