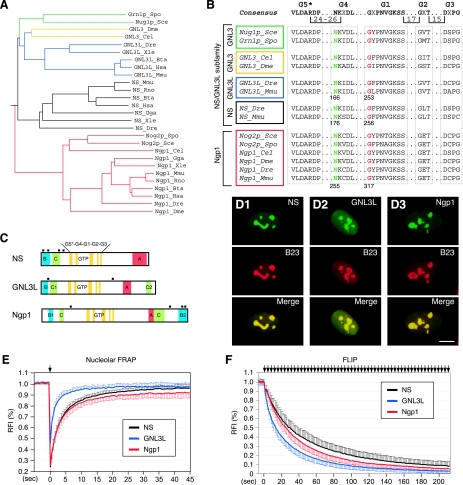
FIG. 1.
NS, GNL3L, and Ngp1 represent a subfamily of GTP-binding proteins with distinct nucleolar localization properties. (A) A rooted phylogenetic tree of NS, GNL3L, and Ngp1 drawn with PHYLIP Drawgram version 3.2. The vertebrate NS genes (black lines) and the GNL3L genes (blue lines) share the same orthologues in the yeast (green lines), fruit fly, and pinworm (yellow lines), whereas the Ngp1 gene (red lines) is the same from yeast to humans. Species abbreviations: Bta, Bos taurus; Cel, Caenorhabditis elegans; Dme, Drosophila melanogaster; Dre, Danio rerio; Gga, Gallus gallus; Hsa, Homo sapiens; Mmu, Mus musculus; Rno, Rattus norvegicus; Sce, Saccharomyces cerevisiae; Spo, Schizosaccharomyces pombe; Xle, Xenopus laevis. (B) The protein sequences of the GTP-binding motifs of the NS family genes were aligned, and the positions of the highly conserved Asn residue (green) in the G4 motif and the Gly residue (red) in the G1 motif were numbered at the bottom. (C) Schematic diagrams of mouse NS, GNL3L, and Ngp1. All genes share an MMR_HSR1 structure, consisting of five circularly permuted GTP-binding motifs (G5*, G4, G1, G2, and G3) and some variations in the B, C, and A domains. Black square boxes indicate nuclear localization signals. (D) The subcellular distributions of NS, GNL3L, and Ngp1 in U2OS cells were revealed by a C-terminally fused GFP (green) and counterstained with anti-B23 immunofluorescence (red). Scale bar, 10 μm. (E) The FRAP recovery rates of NS, GNL3L, and Ngp1 were determined in the nucleoli of U2OS cells transfected with their respective GFP fusion constructs. (F) The FLIP rates of NS, GNL3L, and Ngp1 were measured in U2OS cells, where a small region in the nucleoplasm was repeatedly bleached and the loss of fluorescence signal in the nearest nucleolus was measured over time (see Materials and Methods and Fig. S1B in the supplemental material). The y axes represent the RFI (see Materials and Methods) in the bleached area (for FRAP) or in the nonbleached nucleolus (for FLIP). Error bars represent the standard deviations and are omitted on one side of the curves for clarity. Arrows indicate the bleaching events.