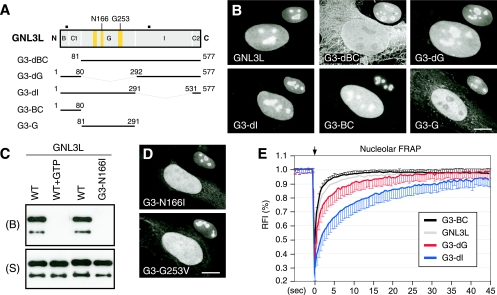
FIG. 2.
The B-C domain of GNL3L is both necessary and sufficient to mediate nucleolar localization. For nucleolar accumulation of the full-length GNL3L, GTP binding is required. (A) Truncated mutants with C-terminally fused GFP were designed to determine the structural requirement for the nucleolar localization of GNL3L. Numbers and black boxes indicate amino acid positions and nuclear localization signals, respectively. Yellow boxes represent, from left to right, the G5*, G1, and G4 motifs. (B) The B-C1 domain is both necessary and sufficient for mediating nucleolar localization. Anti-B23 immunofluorescence of the same cell at a 60% scale is shown in the upper right quadrant of each cell. (C) Wild-type GNL3L (WT) can be retained by GTP-conjugated agarose. The GTP binding of GNL3L is blocked by preincubating GNL3L with 10 mM free GTP (WT+GTP) or by mutating the Asn166 residue in the G4 domain to Ile (mutant G3-N166I). B, bound fraction; S, supernatant. (D) Mutating the Asn166 residue to Ile (mutant G3-N166I) or the Gly253 residue to Val (mutant G3-G253V) perturbs the nucleolar distribution of GNL3L. Anti-B23 staining is shown in the upper right quadrant of each panel. (E) The nucleolar FRAP curves depict the averages of the RFI results in the bleached area relative to the prebleach intensity (set at 1; n = 20) over a 45-s period following photobleaching. Error bars showing standard deviations are omitted on one side for clarity. The FRAP recovery rate of the wild-type protein (GNL3L) mostly resembles the recovery rate of the B-C domain (mutant G3-BC). The arrow indicates the bleach pulse. Scale bars in panels B and D show 10 μm.