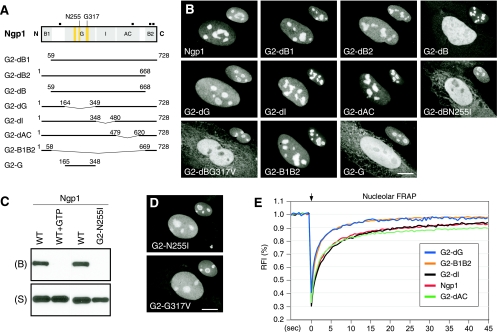
FIG. 3.
The B domains of Ngp1 mediate its nucleolar accumulation, and the G domain controls its nucleolar retention time. (A) C-terminally GFP-fused mutants of Ngp1 were constructed to map the protein domains involved in its static and dynamic distributions. Numbers and black boxes indicate amino acid positions and nuclear localization signals, respectively. Bent line segments indicate deleted protein regions. (B) Distribution analyses showed that deleting both the B1 and B2 domains (mutant G2-dB) disrupts the nucleolar accumulation of Ngp1. The B1-B2 deletion plus a mutation on the Asn255 residue in the G4 motif (mutant G2-dBN255I) or on the Gly317 residue in the G1 motif (mutant G2-dBG317V) completely exclude the protein from the nucleolus. The B1 plus B2 domain (mutant G2-B1B2), but not the G domain (mutant G2-G), is sufficient for accumulation in the nucleolus. Anti-B23 immunofluorescence of the same cells at a 60% scale is shown in the upper right quadrant of each panel. (C) Ngp1 can be retained by GTP-conjugated agarose. The GTP binding of Ngp1 is abolished by preincubating Ngp1 with 10 mM free GTP (WT+GTP) or by mutating the Asn255 residue to Ile (mutant G2-N255I). B, bound fraction; S, supernatant. (D) A single-residue mutation on Asn255 or Gly317 partially perturbs the nucleolar accumulation of Ngp1. (E) Nucleolar FRAP experiments showed that while the B1-B2 domain is necessary and sufficient for mediating nucleolar distribution, it is not enough to recapitulate the dynamic property of the full-length Ngp1 (mutant G2-B1B2). It is the non-nucleolus-targeting G domain that contributes to the retention property of Ngp1 (mutant G2-dG). Deleting the I or AC domain does not alter the dynamic property of Ngp1. Scale bars in panels B and D show 10 μm.