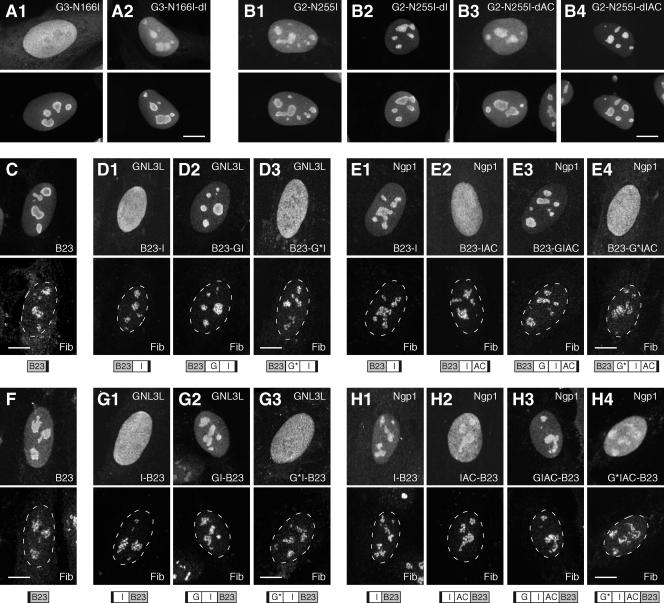
FIG. 4.
Nucleolar accumulation of GNL3L and Ngp1 are controlled by a G-domain-regulated NpLS in the I or IAC domain. (A and B) The nucleolar localization of the G3-N166I (A1) and G2-N255I (B1) mutants can be restored by a deletion of the I domain of GNL3L (A2) or the I or IAC domain of Ngp1 (B2 and B4), but not by a deletion of the AC domain of Ngp1 (B3). The distributions of the mutant proteins were detected by using a C-terminally fused GFP (top panels) and counterstained with anti-B23 immunofluorescence (bottom panels). (C to H) The NpLS activity of the I and IAC domains of GNL3L and Ngp1 remains functional when they are fused to another nucleolar protein, B23. The distribution patterns of B23 (C and F) and B23 fusion proteins (D, E, G, and H) were detected by using a C-terminal HA tag (C to E, upper panels) or an N-terminal Myc tag (F to H, upper panels) and double labeled with antifibrillarin immunofluorescence (Fib, lower panels). The I domain of GNL3L (D1 and G1) and the IAC domain of Ngp1 (E2 and H2), but not the I domain of Ngp1 (E1 and H1), can reduce the nucleolar accumulation of B23. The NpLS activities of the I domain of GNL3L and the IAC domain of Ngp1 can be neutralized by their respective wild-type G domains (D2, E3, G2, and H3), but not by the mutant G domain (G*) with the N166I mutation (for GNL3L; D3 and G3) or the N255I mutation (for Ngp1; E4 and H4). Dashed lines demarcate the nucleocytoplasmic boundaries. The designs of the fusion constructs are depicted below the panels. Black boxes indicate the HA or Myc epitope. Scale bars, 10 μm.