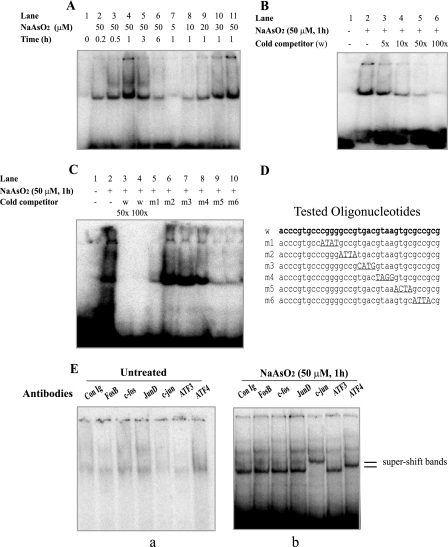
FIG. 4.
Identification of arsenite-activated DNA-binding activity using an EMSA. (A) Arsenite treatment of 10T½ cells activates in vitro protein binding for the 123-bp APE1 promoter fragment. The cells were treated without arsenite or with the indicated arsenite concentration for 30 min, after which the cells were washed and incubated in fresh medium for the time shown. Preparation of cell extracts and EMSA reactions were carried out as described in Materials and Methods. (B) Competition with excess (5- to 100-fold) concentrations of unlabeled wild-type DNA sequence (w). The presence (+) or absence (−) of sodium arsenite or cold competitor is shown. (C) Competition with oligonucleotides containing altered factor binding sites. Competitor oligonucleotides were used at a 50-fold (wild type [w] only) or 100-fold molar excess. Following electrophoresis, the gels were autoradiographed for 16 h. (D) Competitor oligonucleotide sequences. The altered sites are underlined. (E) EMSA supershift only by ATF4 or c-Jun antibodies. Extracts prepared from 10T½ cells treated for 30 min with or without 50 μM sodium arsenite were used in EMSA reactions. Supershift analysis using rabbit immunoglobulin G control (Con Ig) or antibodies directed against the indicated transcription factors was carried out as described in Materials and Methods. The gels were autoradiographed for 16 to 48 h. Only the top portion of the autoradiograms is shown. Separate panels are shown for control (a) and arsenite-treated cells (b).