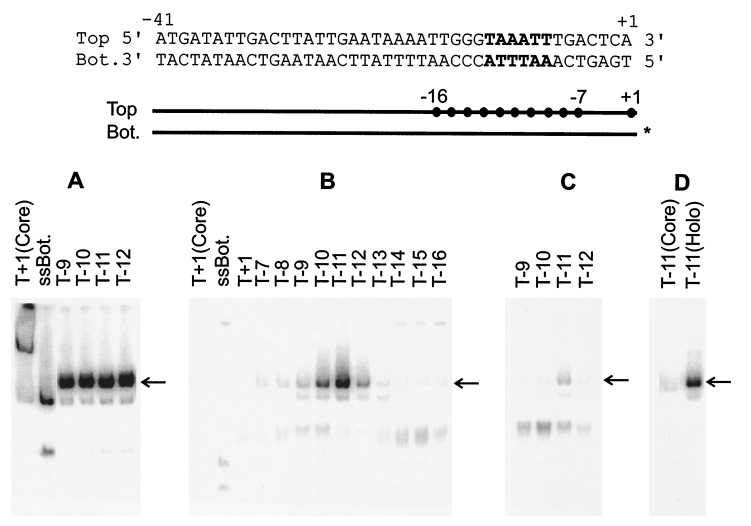
Figure 1.
EMSA of E. coli RNAP holoenzyme with bottom single-stranded fork structures. (Top) The sequence of the parent probe that is derived from the λ late gene promoter PR′, The consensus −10 element is in bold. (Middle) Schematic of the various fork probes with • representing the terminal bases on the top strand used in various probes. The identical bottom strand was annealed to each indicated top strand probe. The 32P- labeled bottom strand is marked with ∗. (Bottom) Results of EMSA using fork probes. In all cases the position of last 3′ base on the top strand (T) is indicated above the appropriate lane. In this and subsequent experiments, the unbound DNA probes were run off the gel. The appropriate shifted band, representing the complex of holoenzyme and DNA fork probe, is marked with an arrow. ssBot refers to a bottom single-stranded probe. Core refers to enzyme-lacking sigma 70. (A) Without heparin. (B) Heparin challenge after holoenzyme incubation with probes. (C) Heparin added before probes. (D) Control as in B.