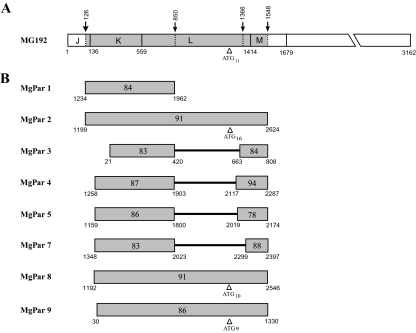
Fig. 1.
Sequence homology between MG192 and MgPars in M. genitalium G37T. A. Schematic representation of the structure of the full-length MG192 gene based on the complete genome sequence of M. genitalium G37T (GenBank Accession No. NC_000908). The nucleotide positions are in relation to the predicted translational start site of the MG192 gene. Divisions J through M indicated by solid vertical lines represent the restriction fragments described previously (Dallo and Baseman, 1991; Peterson et al., 1995). The region highlighted in grey (nt 126–1548) represents the variable region with homology to eight of the nine MgPar sequences shown in (B). This variable region can be further divided into three segments JKL, L and LM (indicated by dotted vertical lines and arrows), which are homologous to different parts of MgPars. Triangle indicates the location of an 11-copy AGT tandem repeat motif (AGT11). B. Schematic representation of the nucleotide sequence alignment of eight MgPars from the M. genitalium G37T genome. Numbers bordering each shaded area refer to the nucleotide positions in the full-length MgPars as described by Iverson-Cabral et al. (2006). Numbers inside the shaded areas indicate the percentage of identity to the corresponding regions in MG192 shown in (A). Triangles indicate the location of the AGT tandem repeat motif with the number of repeat units shown in subscript. Solid black lines in MgPars 3, 4, 5 and 7 represent regions with homology to MG191.