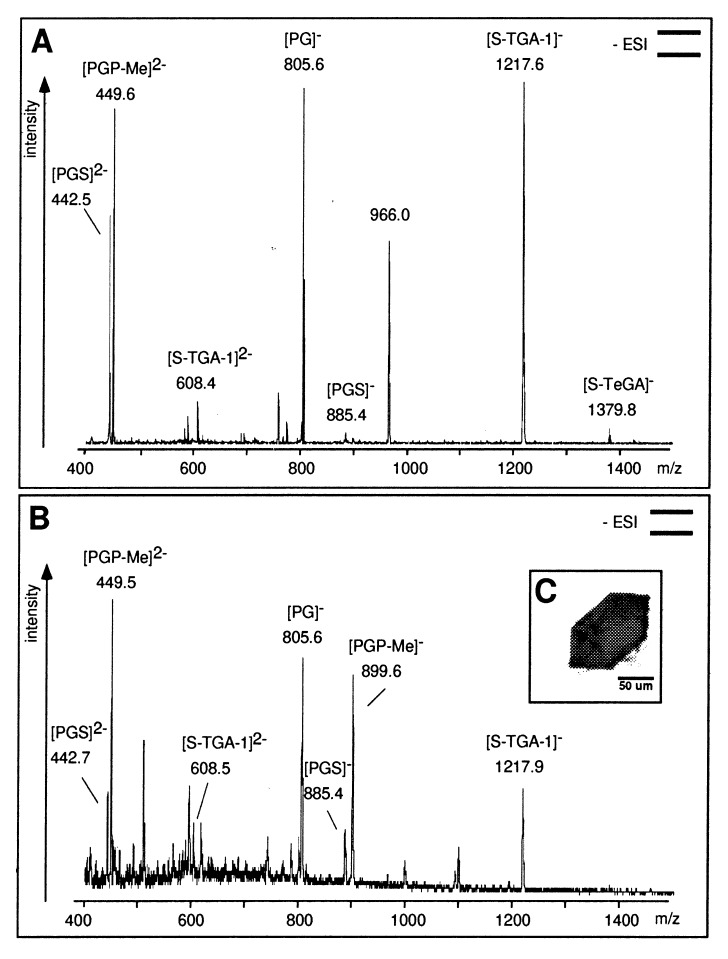
Figure 1.
(A) Negative-ion nanoESI-MS of lipid extracts from PM. The phospholipids PGS and PGP-Me show signals at m/z 442.5, 449.5 (z = 2), and 885.6. The signal at m/z 899.5 is absent because of the double-negative state of detected PGP-Me. PG, the major hydrolysis product of PGS and PGP-Me, was detected at m/z 805.6; phosphatidic acid (PA, m/z 731.5) is absent. The sulfated glycolipids S-TGA-1 and S-TeGA (sulfated tetraglycosyldiphytanylglycerol, a minor species in PM) show signals at m/z 1217.6 and 1379.8. The phospho- and glycolipids were identified by class-specific fragmentations in tandem MS (MS/MS) analyses at their optimized collision energies. (B) NanoESI-MS of the lipid extract of the BR crystal used for crystallographic data collection (shown in C). Pictograms summarize the experimental setup according to ref. 41.