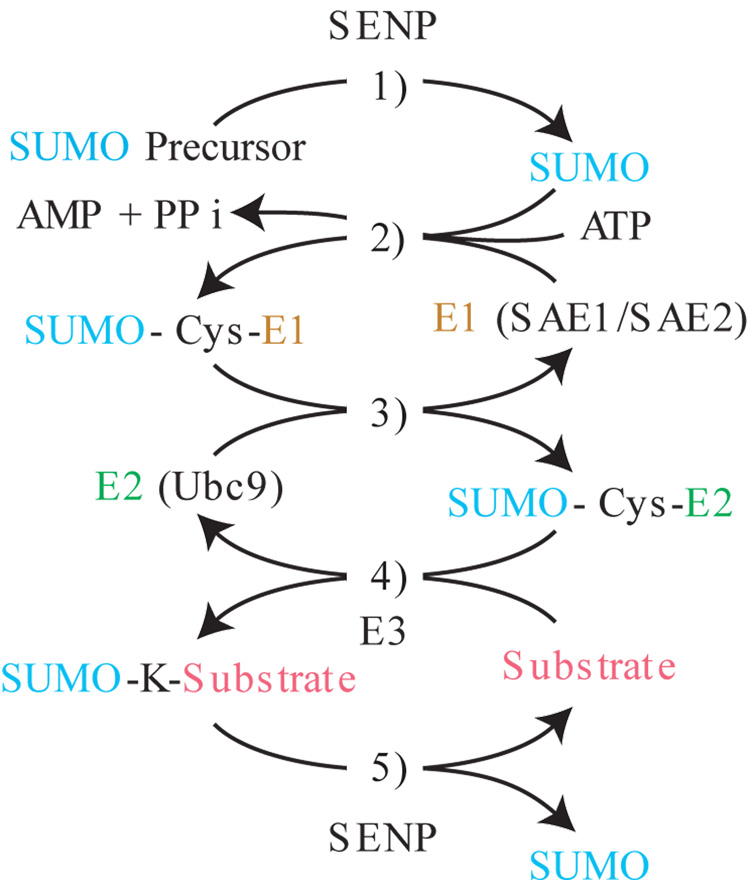
Figure 1. SUMOylation pathway.
1) Initial processing of SUMO precursor by SUMO specific proteases (SENP) removes C terminal residues to generate a new GlyGly C terminus. 2) ATP dependent activation of SUMO by the SUMO specific E1 leads to the formation of a thioester bond between the C terminus of SUMO and a cysteine in the SAE2 subunit. 3) The SUMO moiety is transferred to the SUMO E2 ligase UBC9 through a trans-esterification reaction. 4) Ubc9 catalyzed conjugation of SUMO to substrate leads to the formation of an isopeptide bond between the C terminus of SUMO and the amino group of the target lysine. This step is enhanced by E3 ligases. 5) SUMO conjugation is reversible through the isopeptidase activity of SUMO-specific proteases.