Figure 1.
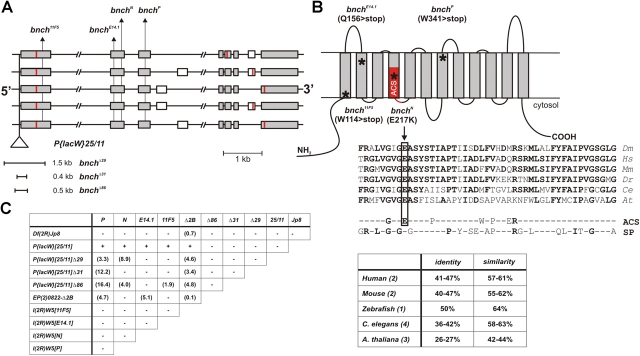
bnch loss of function alleles are semi-lethal and disrupt a highly conserved predicted anion/sugar permease of the MFS. (A) The genomic bnch locus (52E, 14 kb) encodes at least five alternatively spliced transcripts. Translation initiation and termination sites are indicated in red. The approximate location of the P{lacW}[25/11] insertion and point mutations (11F5, E14.1, N, P) is shown. The extent of small deletions (Δ29, Δ31, Δ86) generated by imprecise P-element excision of P{LacW}[25/11] is indicated. The alleles affect gene regions that are common to all alternatively spliced isoforms. (B) Predicted membrane topology for Bnch showing the 12 transmembrane domains typical for proteins of the MFS. The position of the anion/cation (ACS) domain signature is indicated in red. EMS mutations result in the introduction of stop codons (for 11F5, E14.1, and P) or a nonconservative amino acid change (for N) and their positions in the protein are indicated by asterisks. The position of the N missense mutation is shown in a partial multiple alignment of eukaryotic Bnch proteins and consensus sequences of the ACS and sugar porter (SP) subfamilies. The percentages identity and similarity of eukaryotic Bnch proteins to Drosophila Bnch are given in the table. Number of identified Bnch homologues per species is between parentheses. (C) Complementation analysis of bnch point mutations (11F5, E14.1, N, P), excision alleles (Δ29, Δ31, Δ86), deletions (Δ2B, Jp8) and P-element insertion (25/11) indicates allelism. The P{LacW}[25/11] chromosome and its derivatives contain an unrelated lethal mutation that is uncovered by the Jp8 deficiency. The frequency of homozygous and transheterozygous escapers (in parentheses) is not absolute and strongly dependent on the genetic background and culture conditions.