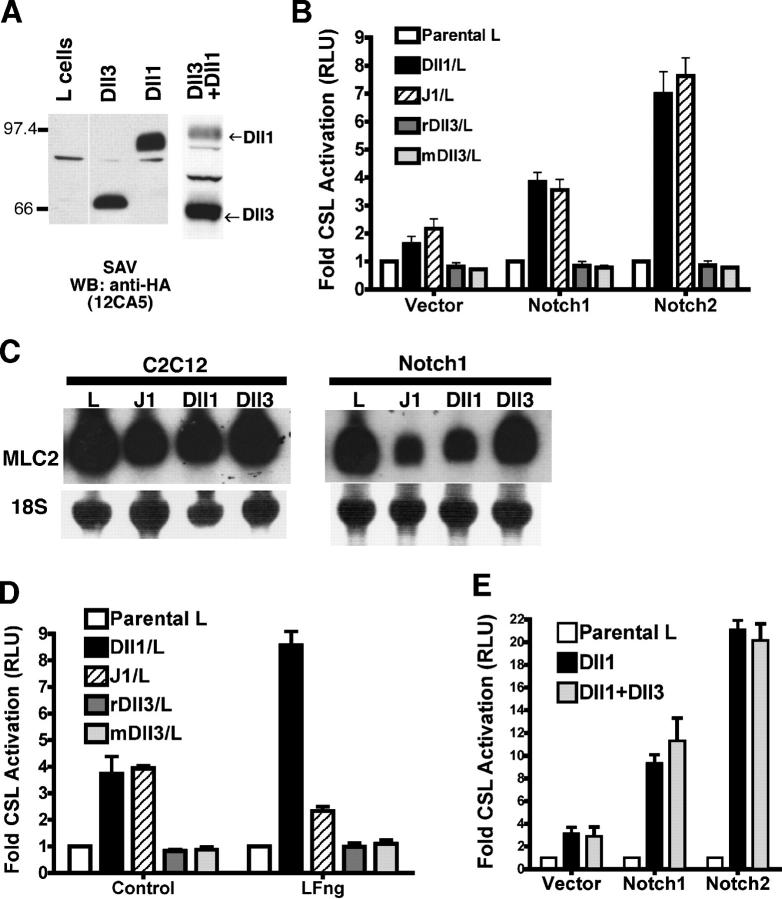
Figure 1.
Dll3 does not activate N signaling in trans. (A) Cell surface expression of HA-tagged Dll1, Dll3, and Dll1 + Dll3 L cell lines was determined by biotinylation, streptavidin (SAV) pull-down, and blotting with anti-HA mAb (12CA5). (B) NIH 3T3 cells transfected with N and CSL reporter were cocultured with J1, Dll1, Dll3, mDll3, or parental (L) cell lines and were assayed for luciferase activity. Error bars reflect the SD of the mean from three experiments. RLU, relative luciferase units. (C) Parental C2C12 and cell lines expressing N1 were cocultured with L, J1, Dll1, or Dll3 cells, and myogenesis was monitored by myosin light chain 2 (MLC2) mRNA expression. Loading and transfer of RNA was monitored by methylene blue staining of 18S rRNA. (D) NIH 3T3 cells transfected with N1, CSL reporter, and control or LFng were cocultured with Dll1, J1, rDll3, mDll3, or L cells and were assayed for luciferase activity (n = 3). (E) Parental L, Dll1, or Dll1 + Dll3 cells were cocultured with NIH 3T3 cells transfected with CSL reporter and N or vector. There was no statistically significant difference between Dll1 and Dll1 + Dll3 (n = 3).