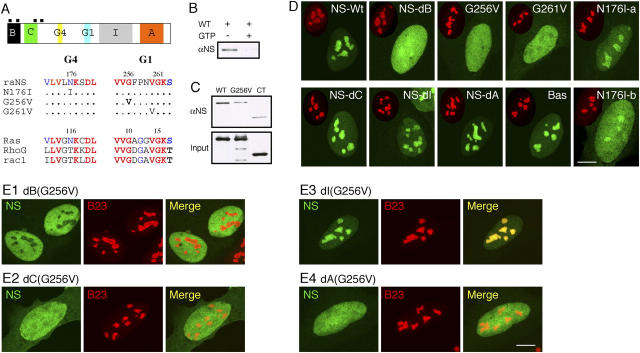
Figure 2.
Nucleolar targeting of NS requires the B-domain and GTP-binding domain, and is blocked by the I-domain. (A) Top: a schematic diagram of the NS protein structure (B: basic; C: coiled-coil; G4, G1: GTP-binding motifs; I: intermediate; A: acidic domains; black boxes: nuclear localization signals). (Bottom) Sequences of single amino acid substitution on the conserved residues in the G4 and G1 motifs. (B) Purified GST fusion protein of wild-type NS could bind GTP-agarose (left lane). This interaction was abolished by competition with 10 mM free GTP (right lane). (C) The role of the G1 motif in mediating the GTP binding of NS was shown by a mutation in the G256 residue (G256V) that reduced its binding affinity to the background level as the COOH-terminal NS control (CT). (D) The static distribution of NS mutants in U2 OS cells (NS-Wt: wild-type; NS-dB: B-domain deletion; NS-dC: C-domain deletion; NS-dI: I-domain deletion; NS-dA: A-domain deletion; Bas: B-domain alone). Green: GFP-fused mutants; red: anti-B23 immunostaining of the same cell (scaled to 60%). (E) Mapping the inhibitory domain that gates the nucleolus-targeting event mediated by the B-domain. A deletion of the I-domain (E3), but not the B- (E1), C- (E2), or A- (E4) domain, can rescue the nucleolar phenotype of G256V mutation. Bars (D and E), 10 μm.