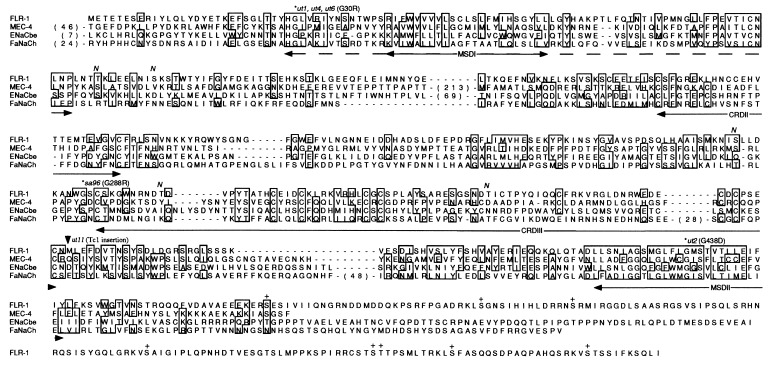
Figure 3.
Sequence alignment of FLR-1 and other ion channels of the DEG/ENaC superfamily. Shown is a comparison of the predicted amino acid sequences between FLR-1, the C. elegans mechanosensory channel MEC-4 (24), the human amiloride-sensitive epithelial sodium channel beta subunit (ENaCbe) (28), and the snail amiloride-sensitive FMRFamide peptide-gated channel (FaNaCh) (30). The residues similar in three or four proteins are boxed. The numbers in parentheses show those of amino acids omitted. Two putative membrane-spanning domains (MSDI and MSDII) and the extracellular Cys-rich domains (CRDII and CRDIII) are shown by lines with arrowheads. A broken line with arrowheads shows the similarity region including MSDI. N and + indicate five potential N-glycosylation sites in the extracellular region and potential phosphorylation sites in the C-terminal intracellular domain of FLR-1, respectively. The latter consist of the consensus sequences for cyclic AMP-dependent protein kinase (R R X S/T), cyclic GMP-dependent protein kinase (R/K R/K X S/T), and Ca2+/calmodulin-dependent protein kinase II (R X X S/T) (32). The mutation sites of (ut1, ut4, ut6), ut2, and sa96 are indicated by ∗ together with the allele names and the resultant amino acid substitution. The Tc1-insertion site in ut11 is indicated by an arrowhead.