Figure 2.
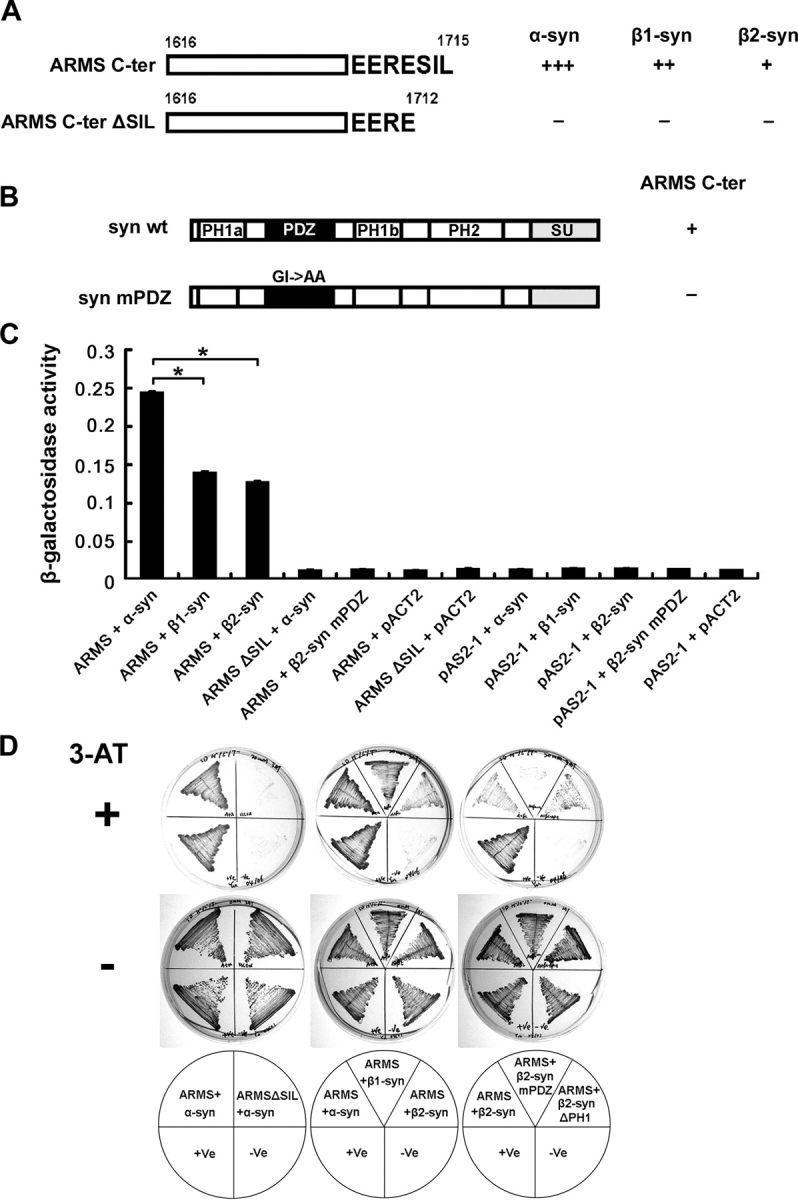
Yeast library screening identified α-syntrophin as an ARMS-interacting protein. (A) α-, β1-, and β2-syntrophins interacted with ARMS COOH terminus (ARMS C-ter, aa 1616–1715) in yeast. Deletion of SIL from the COOH terminus (ARMS C-ter ΔSIL, aa 1616–1712) abolished the protein interaction. +++, very strong; ++, strong; +, moderate; −, no interaction. (B) α-, β1-, and β2-syntrophins are composed of two pleckstrin homology domains (PH1, which is split into PH1a and PH1b by PDZ domain, and PH2), one PDZ domain, and one syntrophin unique domain (SU). Mutation of GLGI loop in syntrophin PDZ domain to GLAA (syn mPDZ) disrupted ARMS–syntrophin interaction. (C) Quantitative β-galactosidase activity assay indicated that α-syntrophins bind to ARMS more strongly than do β-syntrophins in yeast. Arbitrary units were used for the y axis to indicate the relative activity. Each data point represents the mean ± SEM; n = 3; *, P < 0.005. (D) Growth analysis of various yeast transformants on His−/Trp−/Leu− selective plates. In the presence of 20 mM 3-amino-1,2,4-triazole (3-AT), only yeast that expressed interacting proteins grew (top). As a control, all yeast transformants grew normally in the absence of the inhibitor (middle). Bottom panel shows the combinations of different constructs that were transformed into the yeast. +Ve (yeast transformed with pTD1-1 and pVA3-1 plasmids) served as a positive control for this yeast two-hybrid system. −Ve (yeast transformed with pTD1-1 and pAS2-1 plasmids) served as a negative control.
