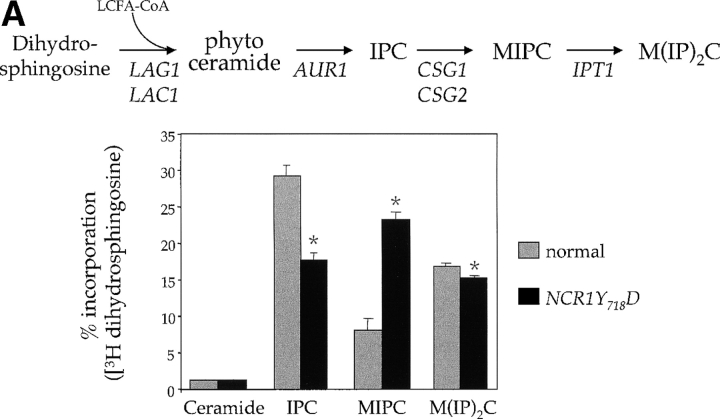
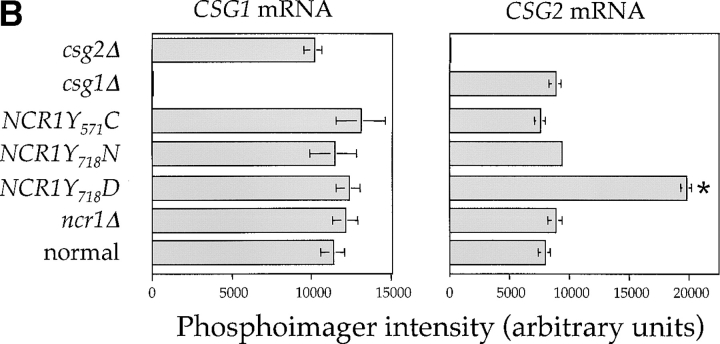
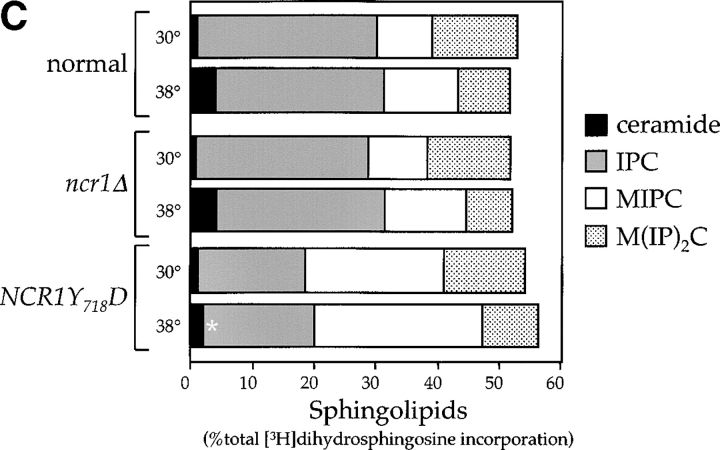
Figure 6.
Sphingolipid metabolism in NCR1 mutants. (A) The sphingolipid biosynthetic pathway and the metabolism of [3H]DHS in normal and NCR1Y 718D cells. Genes for the biosynthesis of ceramide, IPC, MIPC, and M(IP)2C are indicated. Long-chain fatty acylCoA (LCFA-CoA) is an essential substrate for the synthesis of ceramide. Sphingolipids were extracted from the indicated strains grown in the presence of [3H]DHS for 2 h and analyzed by TLC and radio-image scanning. The percent incorporation into all lipids are shown for sphingolipids. Means ± SEM; *, P < 0.01, relative to normal cells. Total incorporation of [3H]DHS for normal and NCR1Y 718D strains was 1.43 and 1.32 × 106 cpm/mg dry weight, respectively. (B) Quantification of RNA hybridization analysis for genes determining the synthesis of MIPC (CSG1 and CSG2). RNA was extracted, resolved, transferred to nitrocellulose membranes by conventional methods, hybridized with CSG1 or CSG2 probes, and analyzed using a phosphorimager (arbitrary units, means ± SEM). Deletion strains csg1Δ and csg2Δ, respectively, for CSG1 and CSG2 acted as hybridization controls. Actin message levels were comparable between strains (not depicted). (C) The stress induction of ceramide synthesis in NCR1 mutants. Cultures of the indicated genotype were grown to exponential phase, split, and grown for a further 2 h in the presence of [3H]DHS at 30 or 38°C. Incorporation of label was normalized to total incorporation into all lipids, which was equivalent at both temperatures (e.g., for normal strains, 2.53 and 2.19 × 106, cpm at 30 and 38°C, respectively). *, P < 0.01.