Figure 9.
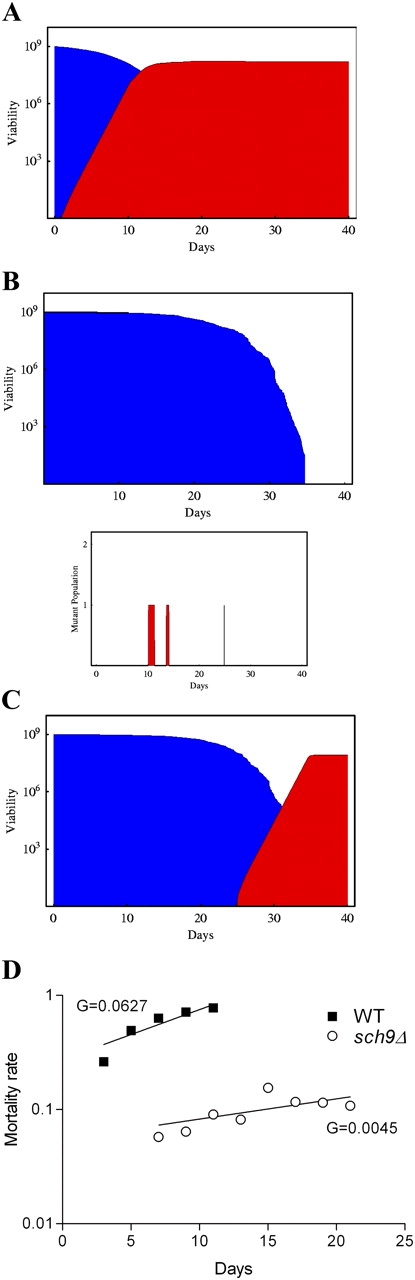
Computational simulation of adaptation to changing environments in S. cerevisiae undergoing programmed or stochastic aging. (A) Results of a computational simulation for programmed aging (model 1) showing the survival of wild-type yeast (blue) and the regrowth of a mutant population (red) that uses the nutrients released by dead cells. Initial population of 109 cells (based on the experimental population size). This result was observed in 52% of the 100 realizations (see text). (B) Results of a computational simulation for stochastic aging (model 2) showing the survival of a long-lived population (blue; sch9Δ) and the regrowth of a mutant population (bottom, red). A typical single realization starting with an initial population of 109 long-lived yeast cells is shown. This result was observed in 99.6% of the 1,000 realizations. The graph on the bottom shows the age-dependent generation/survival of regrowth mutants in sch9Δ cultures. (C) Simulation result for model 2 (B) showing the late regrowth that occurred in 0.4% of the realizations (red). (D) Slopes of the mortality rates (G) for wild-type cells or long-lived mutants (smoothed by using 6-d windows; i.e., mortality at day 5 is the average of the death rates between days 3–5, 5–7, and 7–9).
