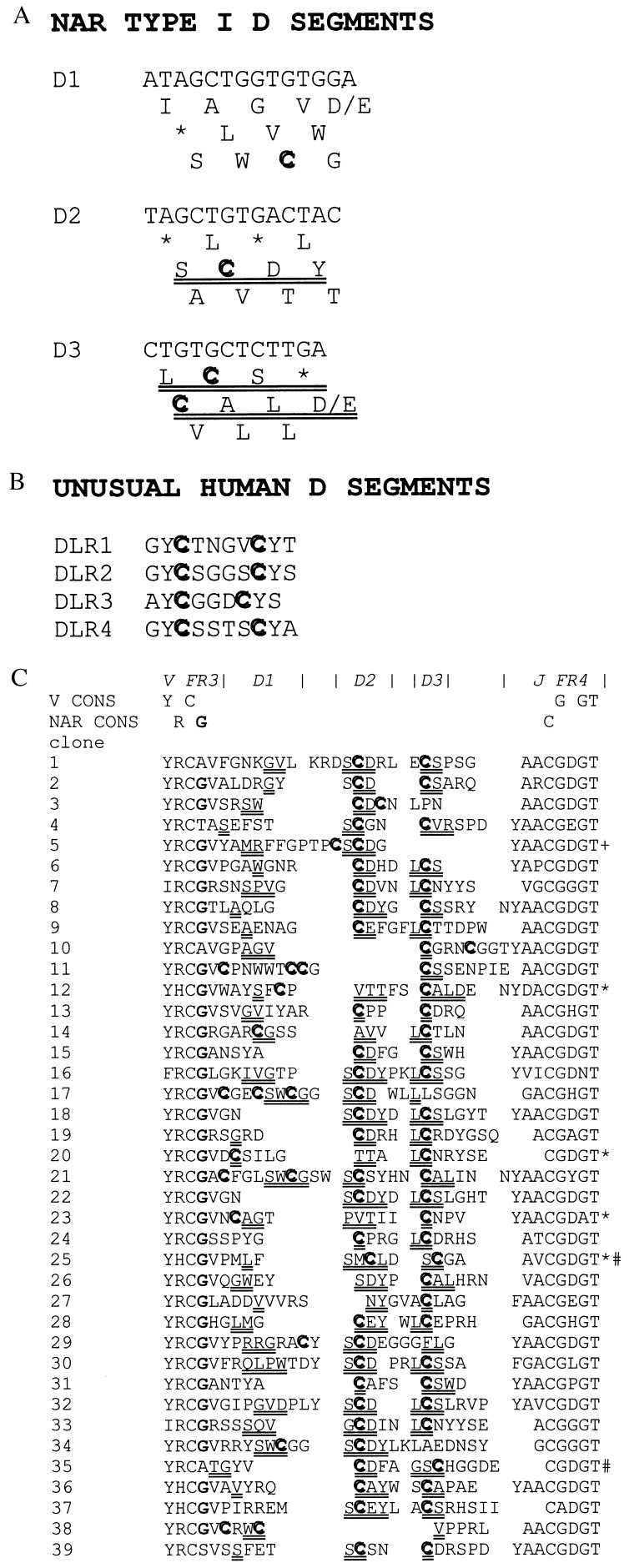
Figure 3.
Nucleotide sequence of NAR Type I D segments. (a) The three reading frames (RF) of D1, D2, and D3. Preferred RF are double underlined, Cys are in bold, and asterisks indicate stop codons. (b) Amino acid sequences of unusual human D segments that form disulfide bridges in some human IgH chains (23). (c) Amino acid sequences deduced from cDNAs of 39 Type I CDR3 junctions in NAR. Amino acids encoded by the D segments are double-underlined (at least two nucleotides of the D region had to be included in the codon encoding the amino acid). Note that sometimes the amino acid encoded by the D cannot be identified in any RF in a presumably because of somatic mutations in the particular cDNA clone or because the first nucleotide of the codon is specified by an N-region addition. In all cases but four (noted by ∗) the D2 segment is “read” in RF “SCDY” (a). D3 is “read” in two RF (“LCS” and “CAL,” a) that encode Cys (# indicates two cases in which D3 encodes Cys, but this was through somatic mutation of nucleotides in D3, not a preferred RF). +, May denote an “oligonucleotide capture” (32) of a D2 segment, i.e., not rearranged in the conventional manner, which is nevertheless “read” in the same RF as most of the other D2 segments. Conserved or invariant residues found in nearly all V domains or are specific only to NAR are indicated above. The NAR-specific glycine (bold, fourth position from left) and Cys in FR4 are displayed on the NAR model in Fig. 4e.