Figure 2.
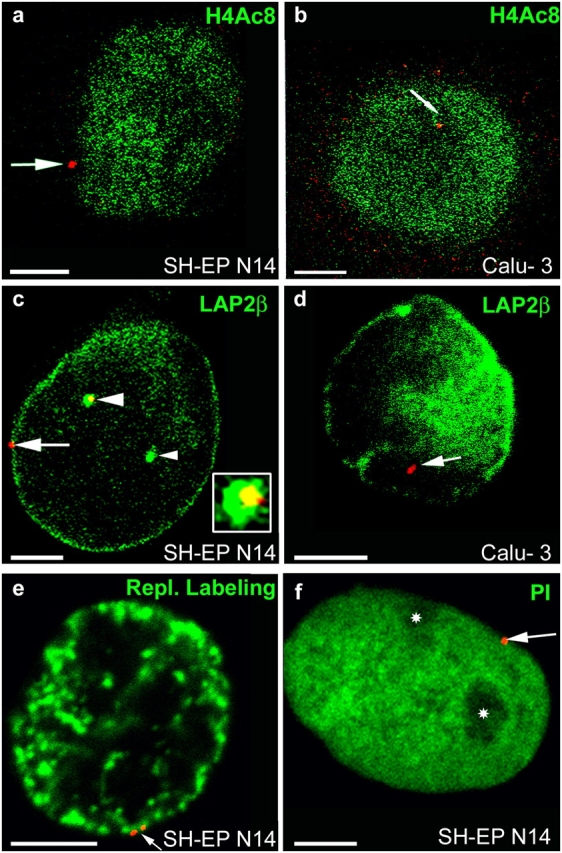
Nuclear localization of CFTR. FISH (red signals) was performed with nuclei from SH-EP N14 (a, c, e, and f) and Calu-3 cells (b and d). Nuclei were immunostained with antibodies against H4Ac8 (a and b, green) or LAP2β (green in c and d). Nuclei were also labeled by replicational pulse labeling (e, green) or were counterstained with propidium iodide (f, green false color). Single light-optical sections are shown and often only one CFTR allele is present in the displayed nuclear plane. (a and b) Arrows point to CFTR alleles associated at the nuclear periphery with chromatin not enriched in H4Ac8 (a, SH-EP N14) or embedded in the nuclear interior into chromatin enriched in H4Ac8 (b, Calu-3). (c) One CFTR allele (arrow) associates at the nuclear periphery with high concentrations of LAP2β. The other CFTR allele (large arrowhead) associates with an invagination of the nuclear periphery into the interior (cross section, green), also containing high concentrations of LAP2β. An enlargement of the FISH signal associated with the invagination is shown in the inset. Only the yellow section of the stained area corresponds to the FISH signal (colocalization of the red FISH signal with the invagination stained in green). The small arrowhead points to a second invagination not associated with a FISH signal. (d) In Calu-3 cells, CFTR (arrow) does not associate with high local concentrations of LAP2β. (e) The replication labeling pattern (green) is typical for the second half of S-phase, with strongly stained perinuclear heterochromatin. The arrow points to a CFTR allele (doublet signal) embedded within the perinuclear heterochromatin. (f) Nucleoli (asterisks) appear unstained in the propidium iodide counterstain (green). The CFTR allele (arrow) is associated with the nuclear periphery, but not with the nucleolar peripheries. Bars, 5 μm.
