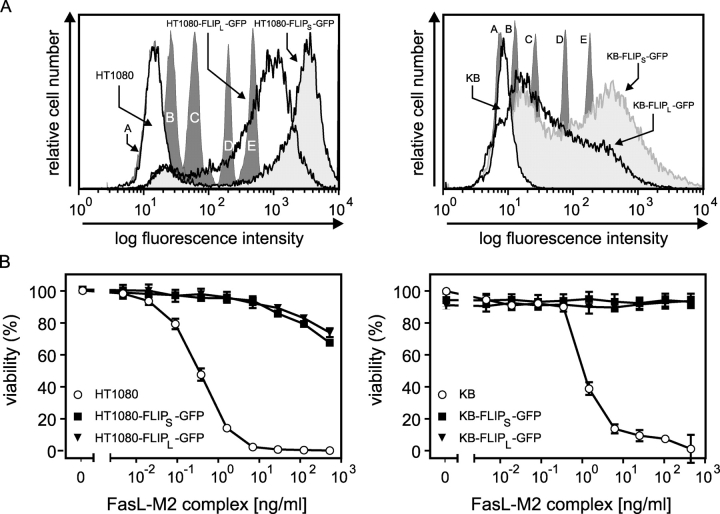
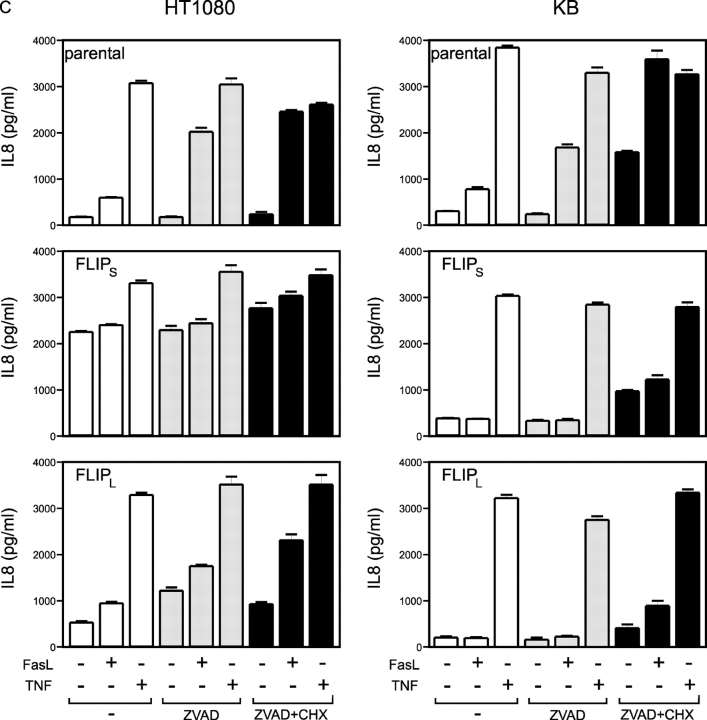
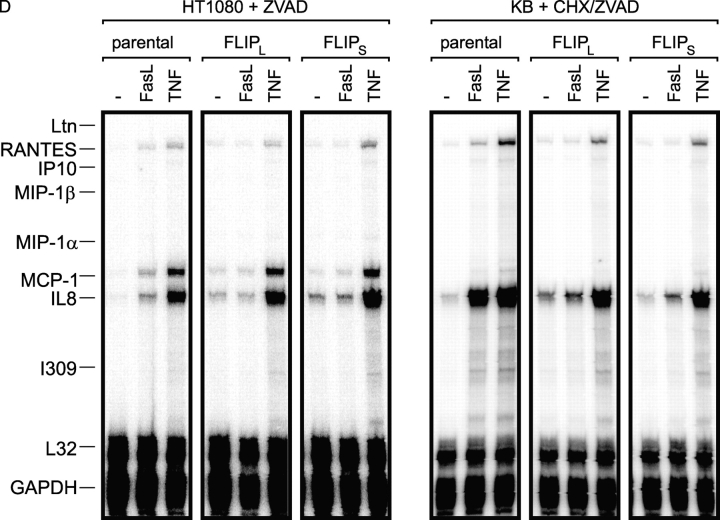
Figure 6.
FLIPL and FLIPS protect HT1080 and KB cells from Fas-mediated apoptosis and block FasL-induced IL8 production. (A) FACS® analysis of GFP FACS® calibration beads with 0 (labeled “A”), 4,500 (labeled “B”), 15,000 (labeled “C”), 44,000 (labeled “D”), and 116,000 (labeled “E”) molecules of equivalent soluble fluorochrome as well as HT1080 and KB cell stably transfected with FLIPL-GFP or FLIPS-GFP. In the case of the KB cells, the localization of the calibration beads was corrected according to a minor difference in the autofluorescence of the parental cells and the nonfluorescent beads. (B) Indicated cells were seeded in 96-well plates (20 × 103 per well) and challenged with the indicated concentrations of soluble Flag-tagged FasL complexed with 0.5 μg/ml of the Flag-specific mAb M2 in the presence of 2.5 μg/ml CHX the next day. After an additional 18 h, cell viability was determined by crystal violet staining. (C) The indicated transfectants and cell lines were seeded in 96-well plates (20 × 103 per well). The next day, the medium was changed and cells were stimulated in triplicates with the indicated combinations of 200 ng/ml of cross-linked Flag-FasL, 20 ng/ml TNF, 20 μM z-VAD-fmk, and 2.5 μg/ml CHX. After 6 h, supernatants were removed and cleared, and IL8 concentrations were determined by ELISA analysis. (D) HT1080 and KB cells and FLIPL-GFP and FLIPS-GFP transfectants derived thereof were challenged for 6 h with 200 ng/ml of M2–cross-linked Flag-FasL or 20 ng/ml TNF in the presence of 20 μM z-VAD-fmk (HT1080) or a mixture of 2.5 μg/ml CHX and 20 μM Z-VAD-FMK (KB). Total RNAs were isolated and the expression of the indicated genes was determined by RPA analysis.